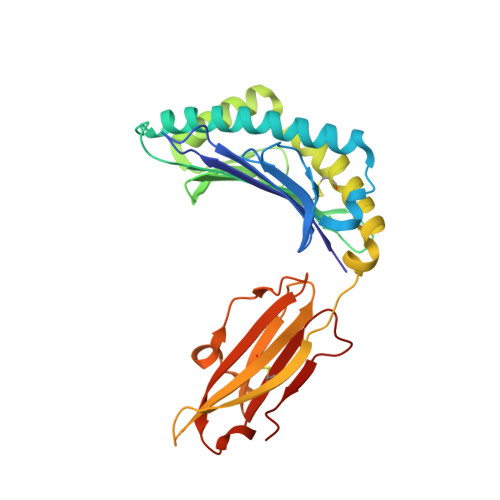
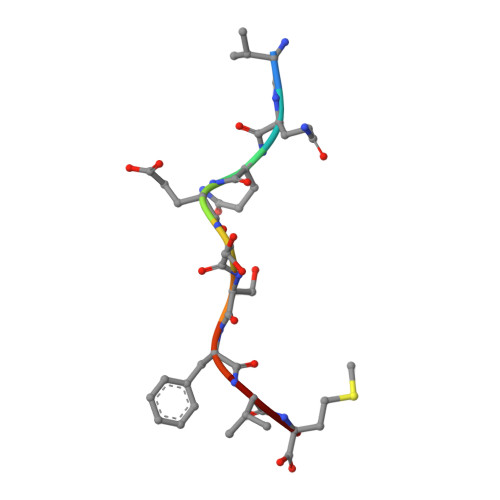
Structure of a SARS coronavirus-derived peptide bound to the human major histocompatibility complex class I molecule HLA-B*1501.
Roder, G., Kristensen, O., Kastrup, J.S., Buus, S., Gajhede, M.(2008) Acta Crystallogr Sect F Struct Biol Cryst Commun 64: 459-462
- PubMed: 18540051
- DOI: https://doi.org/10.1107/S1744309108012396
- Primary Citation of Related Structures:
3C9N - PubMed Abstract:
The human leukocyte antigen (HLA) class I system comprises a highly polymorphic set of molecules that specifically bind and present peptides to cytotoxic T cells. HLA-B*1501 is a prototypical member of the HLA-B62 supertype and only two peptide-HLA-B*1501 structures have been determined. Here, the crystal structure of HLA-B*1501 in complex with a SARS coronavirus-derived nonapeptide (VQQESSFVM) has been determined at high resolution (1.87 A). The peptide is deeply anchored in the B and F pockets, but with the Glu4 residue pointing away from the floor in the peptide-binding groove, making it available for interactions with a potential T-cell receptor.
- Institute of International Health, Immunology and Microbiology, University of Copenhagen, Blegdamsvej 3, DK-2200 Copenhagen, Denmark. g.roder@immi.ku.dk
Organizational Affiliation: