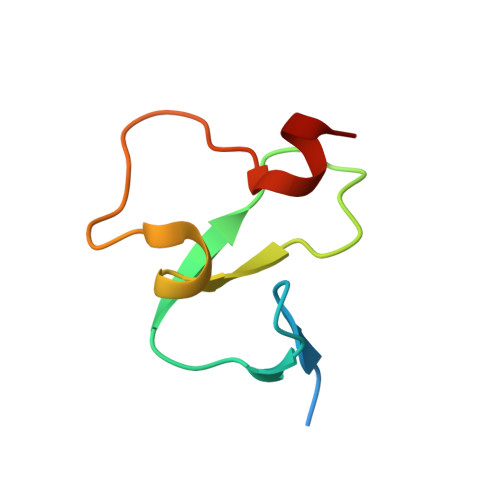
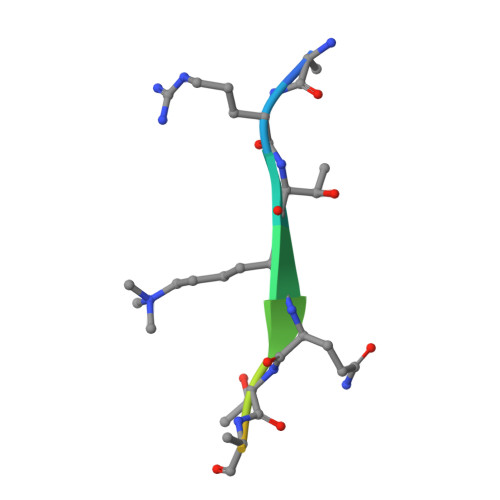
The crystal structure of the ING5 PHD finger in complex with an H3K4me3 histone peptide.
Champagne, K.S., Saksouk, N., Pena, P.V., Johnson, K., Ullah, M., Yang, X.J., Cote, J., Kutateladze, T.G.(2008) Proteins 72: 1371-1376
- PubMed: 18623064
- DOI: https://doi.org/10.1002/prot.22140
- Primary Citation of Related Structures:
3C6W - Department of Pharmacology, University of Colorado at Denver, Aurora, Colorado 80045, USA. karen.champagne@uchsc.edu
Organizational Affiliation: