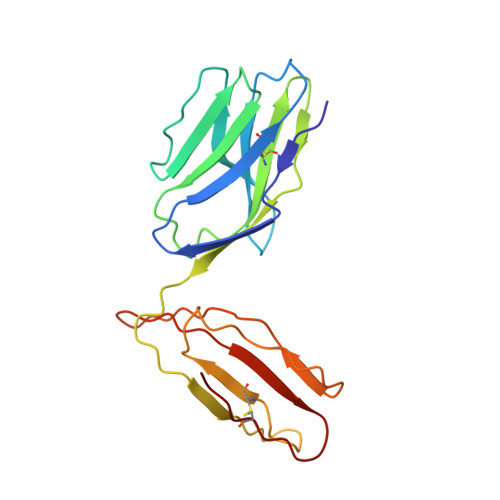
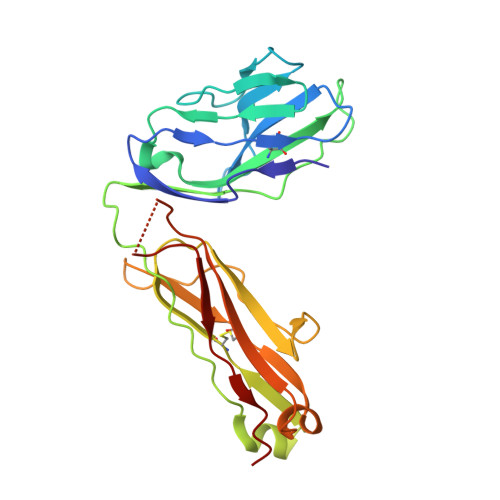
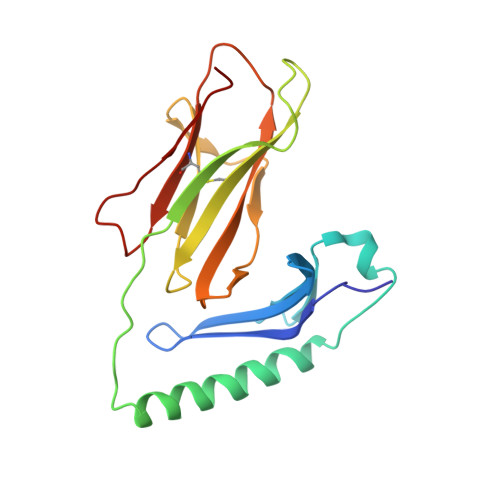
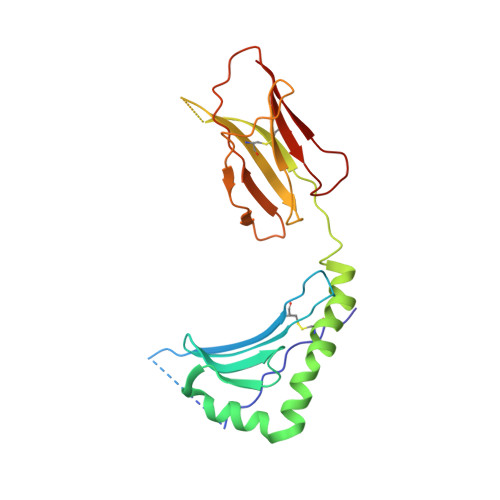
Crossreactive T Cells spotlight the germline rules for alphabeta T cell-receptor interactions with MHC molecules.
Dai, S., Huseby, E.S., Rubtsova, K., Scott-Browne, J., Crawford, F., Macdonald, W.A., Marrack, P., Kappler, J.W.(2008) Immunity 28: 324-334
- PubMed: 18308592
- DOI: https://doi.org/10.1016/j.immuni.2008.01.008
- Primary Citation of Related Structures:
3C5Z, 3C60, 3C6L - PubMed Abstract:
To test whether highly crossreactive alphabeta T cell receptors (TCRs) produced during limited negative selection best illustrate evolutionarily conserved interactions between TCR and major histocompatibility complex (MHC) molecules, we solved the structures of three TCRs bound to the same MHC II peptide (IAb-3K). The TCRs had similar affinities for IAb-3K but varied from noncrossreactive to extremely crossreactive with other peptides and MHCs. Crossreactivity correlated with a shrinking, increasingly hydrophobic TCR-ligand interface, involving fewer TCR amino acids. A few CDR1 and CDR2 amino acids dominated the most crossreactive TCR interface with MHC, including Vbeta8 48Y and 54E and Valpha4 29Y, arranged to impose the familiar diagonal orientation of TCR on MHC. These interactions contribute to MHC binding by other TCRs using related V regions, but not usually so dominantly. These data show that crossreactive TCRs can spotlight the evolutionarily conserved features of TCR-MHC interactions and that these interactions impose the diagonal docking of TCRs on MHC.
- Howard Hughes Medical Institute and National Jewish Medical and Research Center, Denver, CO 80206, USA.
Organizational Affiliation: