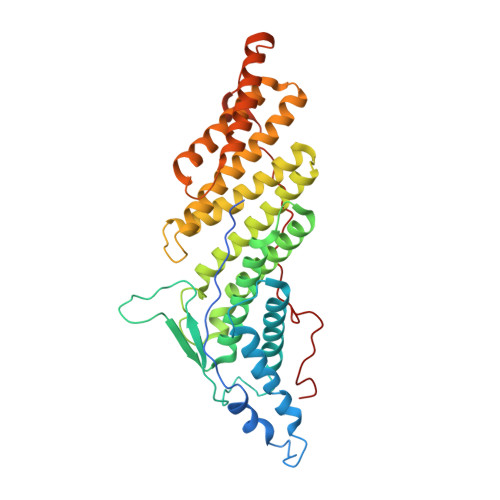
ALIX-CHMP4 interactions in the human ESCRT pathway.
McCullough, J., Fisher, R.D., Whitby, F.G., Sundquist, W.I., Hill, C.P.(2008) Proc Natl Acad Sci U S A 105: 7687-7691
- PubMed: 18511562
- DOI: https://doi.org/10.1073/pnas.0801567105
- Primary Citation of Related Structures:
3C3O, 3C3Q, 3C3R - PubMed Abstract:
The ESCRT pathway facilitates membrane fission events during enveloped virus budding, multivesicular body formation, and cytokinesis. To promote HIV budding and cytokinesis, the ALIX protein must bind and recruit CHMP4 subunits of the ESCRT-III complex, which in turn participate in essential membrane remodeling functions. Here, we report that the Bro1 domain of ALIX binds specifically to C-terminal residues of the human CHMP4 proteins (CHMP4A-C). Crystal structures of the complexes reveal that the CHMP4 C-terminal peptides form amphipathic helices that bind across the conserved concave surface of ALIX(Bro1). ALIX-dependent HIV-1 budding is blocked by mutations in exposed ALIX(Bro1) residues that help contribute to the binding sites for three essential hydrophobic residues that are displayed on one side of the CHMP4 recognition helix (M/L/IxxLxxW). The homologous CHMP1-3 classes of ESCRT-III proteins also have C-terminal amphipathic helices, but, in those cases, the three hydrophobic residues are arrayed with L/I/MxxxLxxL spacing. Thus, the distinct patterns of hydrophobic residues provide a "code" that allows the different ESCRT-III subunits to bind different ESCRT pathway partners, with CHMP1-3 proteins binding MIT domain-containing proteins, such as VPS4 and Vta1/LIP5, and CHMP4 proteins binding Bro1 domain-containing proteins, such as ALIX.
- Department of Biochemistry, University of Utah, Salt Lake City, UT 84112-5650, USA.
Organizational Affiliation: