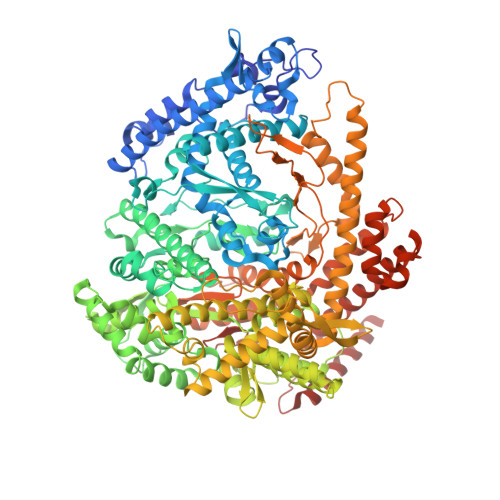
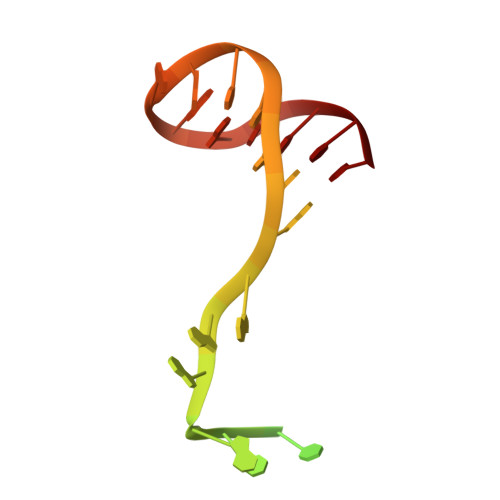Structural basis for DNA-hairpin promoter recognition by the bacteriophage N4 virion RNA polymerase.
Gleghorn, M.L., Davydova, E.K., Rothman-Denes, L.B., Murakami, K.S.(2008) Mol Cell 32: 707-717
- PubMed: 19061645
- DOI: https://doi.org/10.1016/j.molcel.2008.11.010
- Primary Citation of Related Structures:
3C2P, 3C3L, 3C46 - PubMed Abstract:
Coliphage N4 virion-encapsidated RNA polymerase (vRNAP) is a member of the phage T7-like single-subunit RNA polymerase (RNAP) family. Its central domain (mini-vRNAP) contains all RNAP functions of the full-length vRNAP, which recognizes a 5 to 7 base pair stem and 3 nucleotide loop hairpin DNA promoter. Here, we report the X-ray crystal structures of mini-vRNAP bound to promoters. Mini-vRNAP uses four structural motifs to recognize DNA sequences at the hairpin loop and stem and to unwind DNA. Despite their low sequence similarity, three out of four motifs are shared with T7 RNAP that recognizes a double-stranded DNA promoter. The binary complex structure and results of engineered disulfide linkage experiments reveal that the plug and motif B loop, which block the access of template DNA to the active site in the apo-form mini-vRNAP, undergo a large-scale conformational change upon promoter binding, explaining the restricted promoter specificity that is critical for N4 phage early transcription.
- Department of Biochemistry and Molecular Biology, The Pennsylvania State University, University Park, PA 16802, USA.
Organizational Affiliation: