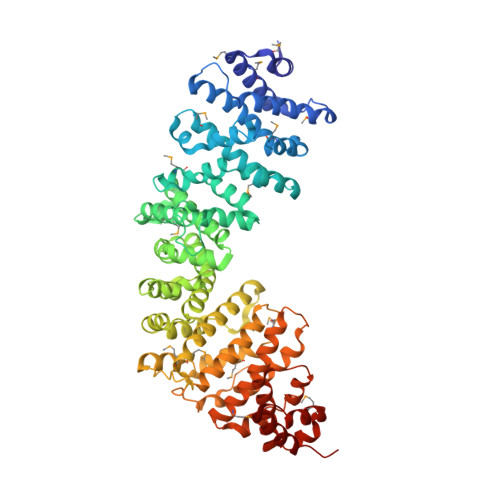
The C. elegans SYS-1 protein is a bona fide beta-catenin.
Liu, J., Phillips, B.T., Amaya, M.F., Kimble, J., Xu, W.(2008) Dev Cell 14: 751-761
- PubMed: 18477457
- DOI: https://doi.org/10.1016/j.devcel.2008.02.015
- Primary Citation of Related Structures:
3C2G, 3C2H - PubMed Abstract:
C. elegans SYS-1 has key functional characteristics of a canonical beta-catenin, but no significant sequence similarity. Here, we report the SYS-1 crystal structure, both on its own and in a complex with POP-1, the C. elegans TCF homolog. The two structures possess signature features of canonical beta-catenin and the beta-catenin/TCF complex that could not be predicted by sequence. Most importantly, SYS-1 bears 12 armadillo repeats and the SYS-1/POP-1 interface is anchored by a conserved salt-bridge, the "charged button." We also modeled structures for three other C. elegans beta-catenins to predict the molecular basis of their distinct binding properties. Finally, we generated a phylogenetic tree, using the region of highest structural similarity between SYS-1 and beta-catenin, and found that SYS-1 clusters robustly within the beta-catenin clade. We conclude that the SYS-1 protein belongs to the beta-catenin family and suggest that additional divergent beta-catenins await discovery.
- Department of Biological Structure, University of Washington, Seattle, WA 98195-7420, USA.
Organizational Affiliation: