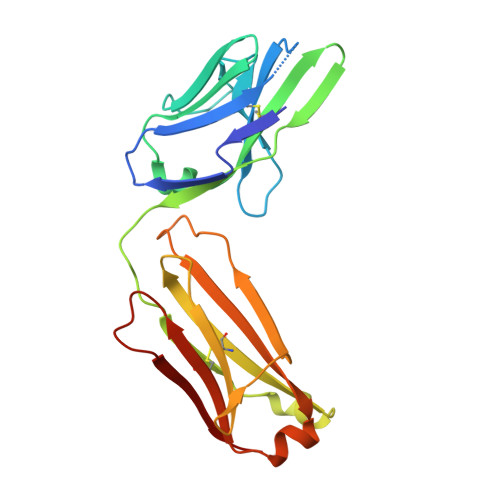
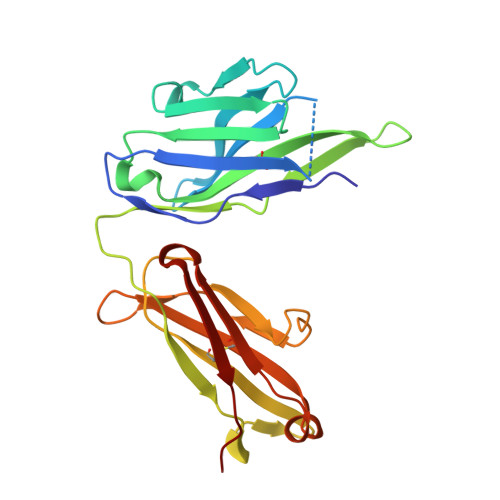
Matuzumab binding to EGFR prevents the conformational rearrangement required for dimerization.
Schmiedel, J., Blaukat, A., Li, S., Knochel, T., Ferguson, K.M.(2008) Cancer Cell 13: 365-373
- PubMed: 18394559
- DOI: https://doi.org/10.1016/j.ccr.2008.02.019
- Primary Citation of Related Structures:
3C08, 3C09 - PubMed Abstract:
An increasing number of therapeutic antibodies targeting tumors that express the epidermal growth factor receptor (EGFR) are in clinical use or late stages of clinical development. Here we investigate the molecular basis for inhibition of EGFR activation by the therapeutic antibody matuzumab (EMD72000). We describe the X-ray crystal structure of the Fab fragment of matuzumab (Fab72000) in complex with isolated domain III from the extracellular region of EGFR. Fab72000 interacts with an epitope on EGFR that is distinct from the ligand-binding region on domain III and from the cetuximab/Erbitux epitope. Matuzumab blocks ligand-induced receptor activation indirectly by sterically preventing the domain rearrangement and local conformational changes that must occur for high-affinity ligand binding and receptor dimerization.
- Department of Physiology, University of Pennsylvania School of Medicine, Philadelphia, PA 19104, USA.
Organizational Affiliation: