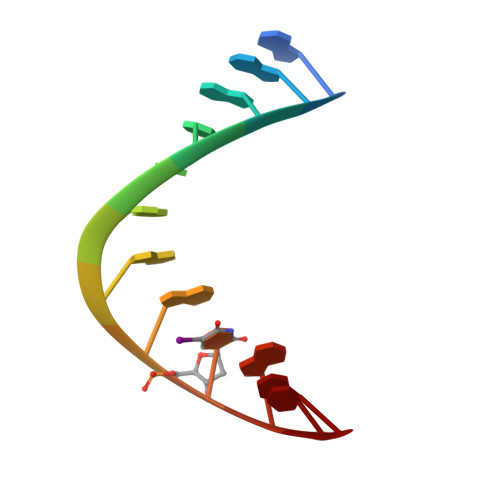
Specific recognition of RNA/DNA hybrid and enhancement of human RNase H1 activity by HBD.
Nowotny, M., Cerritelli, S.M., Ghirlando, R., Gaidamakov, S.A., Crouch, R.J., Yang, W.(2008) EMBO J 27: 1172-1181
- PubMed: 18337749
- DOI: https://doi.org/10.1038/emboj.2008.44
- Primary Citation of Related Structures:
3BSU - PubMed Abstract:
Human RNase H1 contains an N-terminal domain known as dsRHbd for binding both dsRNA and RNA/DNA hybrid. We find that dsRHbd binds preferentially to RNA/DNA hybrids by over 25-fold and rename it as hybrid binding domain (HBD). The crystal structure of HBD complexed with a 12 bp RNA/DNA hybrid reveals that the RNA strand is recognized by a protein loop, which forms hydrogen bonds with the 2'-OH groups. The DNA interface is highly specific and contains polar residues that interact with the phosphate groups and an aromatic patch that appears selective for binding deoxyriboses. HBD is unique relative to non-sequence-specific dsDNA- and dsRNA-binding domains because it does not use positive dipoles of alpha-helices for nucleic acid binding. Characterization of full-length enzymes with defective HBDs indicates that this domain dramatically enhances both the specific activity and processivity of RNase H1. Similar activity enhancement by small substrate-binding domains linked to the catalytic domain likely occurs in other nucleic acid enzymes.
- Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, MD 20892, USA.
Organizational Affiliation: