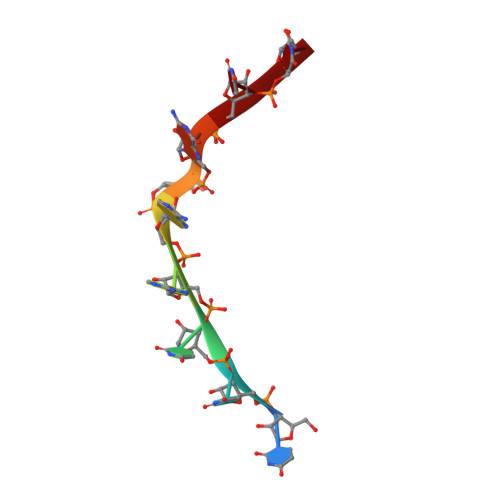
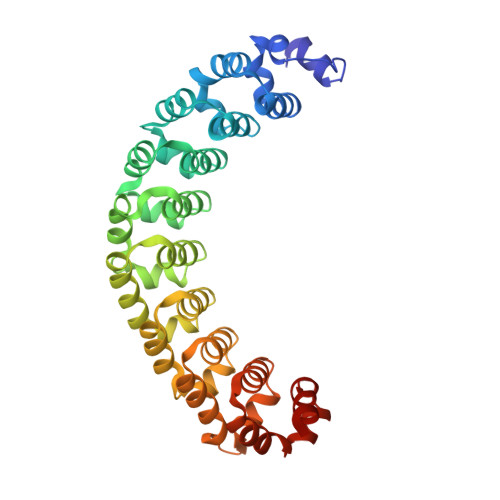Structures of human Pumilio with noncognate RNAs reveal molecular mechanisms for binding promiscuity.
Gupta, Y.K., Nair, D.T., Wharton, R.P., Aggarwal, A.K.(2008) Structure 16: 549-557
- PubMed: 18328718
- DOI: https://doi.org/10.1016/j.str.2008.01.006
- Primary Citation of Related Structures:
3BSB, 3BSX - PubMed Abstract:
Pumilio is a founder member of the evolutionarily conserved Puf family of RNA-binding proteins that control a number of physiological processes in eukaryotes. A structure of human Pumilio (hPum) Puf domain bound to a Drosophila regulatory sequence showed that each Puf repeat recognizes a single nucleotide. Puf domains in general bind promiscuously to a large set of degenerate sequences, but the structural basis for this promiscuity has been unclear. Here, we describe the structures of hPum Puf domain complexed to two noncognate RNAs, CycB(reverse) and Puf5. In each complex, one of the nucleotides is ejected from the binding surface, in effect, acting as a "spacer." The complexes also reveal the plasticity of several Puf repeats, which recognize noncanonical nucleotides. Together, these complexes provide a molecular basis for recognition of degenerate binding sites, which significantly increases the number of mRNAs targeted for regulation by Puf proteins in vivo.
- Department of Structural and Chemical Biology, Mount Sinai School of Medicine, Box 1677, 1425 Madison Avenue, New York, NY 10029, USA.
Organizational Affiliation: