The Interplay of Ligand Binding and Phosphorylation in the Regulation of Dynein Light Chain LC8
Benison, G.C., Chiodo, M., Karplus, P.A., Barbar, E.J.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Dynein light chain 1, cytoplasmic | 89 | Drosophila melanogaster | Mutation(s): 1 | 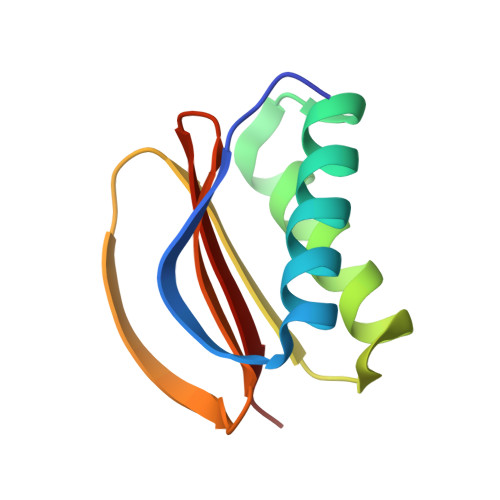 | |
UniProt | |||||
Find proteins for Q24117 (Drosophila melanogaster) Explore Q24117 Go to UniProtKB: Q24117 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q24117 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Protein swallow 10-resiude peptide | B [auth C] | 10 | N/A | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P40688 (Drosophila melanogaster) Explore P40688 Go to UniProtKB: P40688 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P40688 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 44.194 | α = 90 |
| b = 44.194 | β = 90 |
| c = 203.218 | γ = 120 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| PDB_EXTRACT | data extraction |