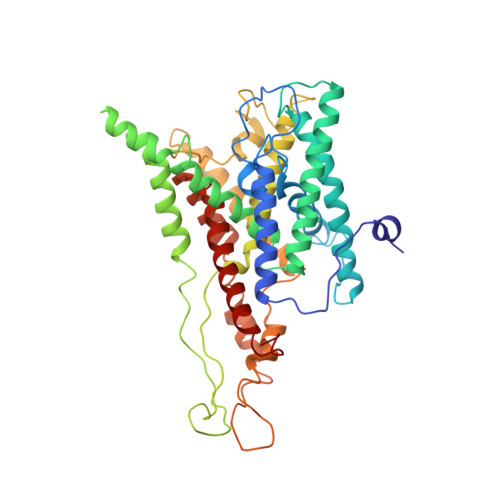
Ribosome binding of a single copy of the SecY complex: implications for protein translocation
Menetret, J.F., Schaletzky, J., Clemons, W.M., Osborne, A.R., Skanland, S.S., Denison, C., Gygi, S.P., Kirkpatrick, D.S., Park, E., Ludtke, S.J., Rapoport, T.A., Akey, C.W.(2007) Mol Cell 28: 1083-1092
- PubMed: 18158904
- DOI: https://doi.org/10.1016/j.molcel.2007.10.034
- Primary Citation of Related Structures:
3BO0, 3BO1 - PubMed Abstract:
The SecY complex associates with the ribosome to form a protein translocation channel in the bacterial plasma membrane. We have used cryo-electron microscopy and quantitative mass spectrometry to show that a nontranslating E. coli ribosome binds to a single SecY complex. The crystal structure of an archaeal SecY complex was then docked into the electron density maps. In the resulting model, two cytoplasmic loops of SecY extend into the exit tunnel near proteins L23, L29, and L24. The loop between transmembrane helices 8 and 9 interacts with helices H59 and H50 in the large subunit RNA, while the 6/7 loop interacts with H7. We also show that point mutations of basic residues within either loop abolish ribosome binding. We suggest that SecY binds to this primary site on the ribosome and subsequently captures and translocates the nascent chain.
- Department of Physiology and Biophysics, Boston University School of Medicine, 700 Albany Street, Boston, MA 02118-2526, USA.
Organizational Affiliation: