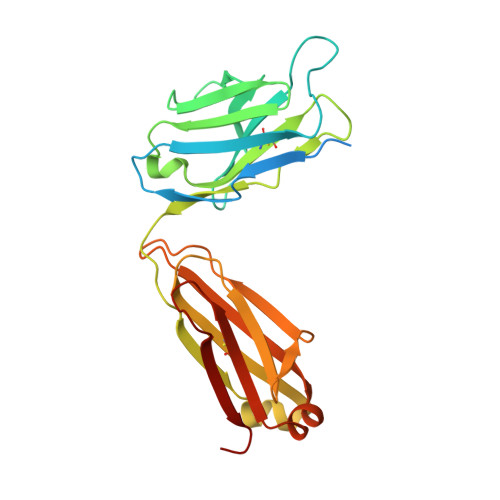
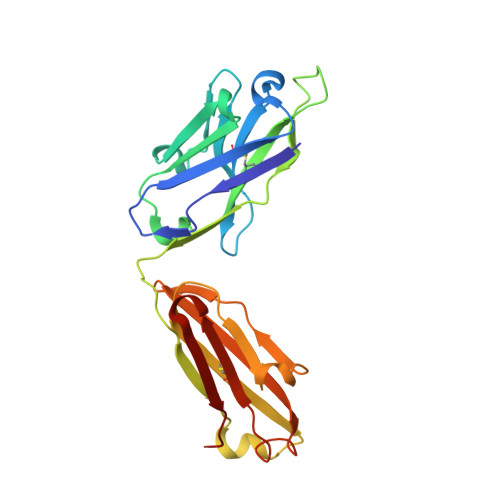
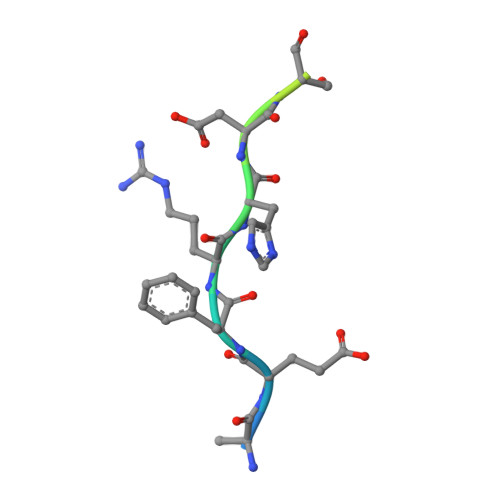
Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope.
Miles, L.A., Wun, K.S., Crespi, G.A., Fodero-Tavoletti, M.T., Galatis, D., Bagley, C.J., Beyreuther, K., Masters, C.L., Cappai, R., McKinstry, W.J., Barnham, K.J., Parker, M.W.(2008) J Mol Biology 377: 181-192
- PubMed: 18237744
- DOI: https://doi.org/10.1016/j.jmb.2007.12.036
- Primary Citation of Related Structures:
3BAE, 3BKC, 3BKJ, 3BKM - PubMed Abstract:
Alzheimer's disease (AD) is the most common form of dementia. Amyloid-beta (A beta) peptide, generated by proteolytic cleavage of the amyloid precursor protein, is central to AD pathogenesis. Most pharmaceutical activity in AD research has focused on A beta, its generation and clearance from the brain. In particular, there is much interest in immunotherapy approaches with a number of anti-A beta antibodies in clinical trials. We have developed a monoclonal antibody, called WO2, which recognises the A beta peptide. To this end, we have determined the three-dimensional structure, to near atomic resolution, of both the antibody and the complex with its antigen, the A beta peptide. The structures reveal the molecular basis for WO2 recognition and binding of A beta. The A beta peptide adopts an extended, coil-like conformation across its major immunodominant B-cell epitope between residues 2 and 8. We have also studied the antibody-bound A beta peptide in the presence of metals known to affect its aggregation state and show that WO2 inhibits these interactions. Thus, antibodies that target the N-terminal region of A beta, such as WO2, hold promise for therapeutic development.
- Biota Structural Biology Laboratory, St. Vincent's Institute of Medical Research, 9 Princes Street, Fitzroy, Victoria 3065, Australia.
Organizational Affiliation: