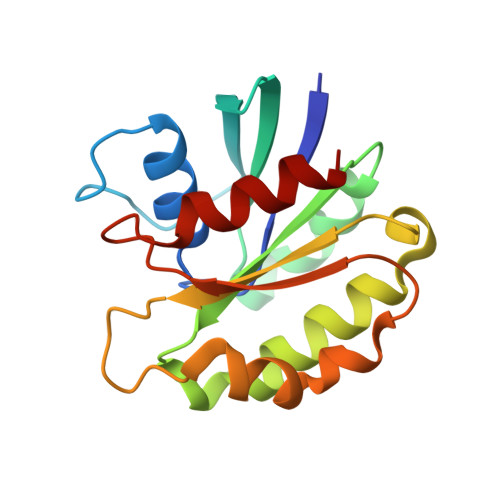
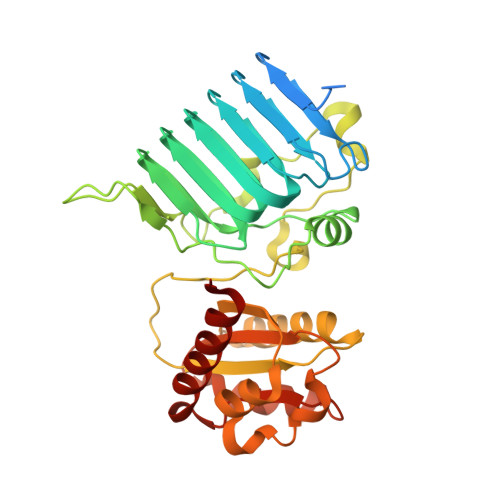
The retinitis pigmentosa 2 gene product is a GTPase-activating protein for Arf-like 3
Veltel, S., Gasper, R., Eisenacher, E., Wittinghofer, A.(2008) Nat Struct Mol Biol 15: 373-380
- PubMed: 18376416
- DOI: https://doi.org/10.1038/nsmb.1396
- Primary Citation of Related Structures:
3BH6, 3BH7 - PubMed Abstract:
The retinitis pigmentosa 2 (RP2) gene is responsible for a particular variant of X chromosome-linked eye disease. Previously, RP2 was shown to bind the GTP form of the small G protein Arf-like 3 (Arl3), thus qualifying as an effector. Here we present the Arl3-GppNHp-RP2 complex structure, which shows features resembling complexes with GTPase-activating proteins (GAPs). Biochemical analysis showing a 90,000-fold stimulation of the GTPase reaction together with the structure of an Arl3-GDP-AlF4--RP2 transition state complex showed that RP2 is an efficient GAP for Arl3, with structural features similar to other GAPs. Furthermore, the effect of mutations in patients with retinitis pigmentosa correlated with their effect on catalysis, in particular the mutation of the arginine finger of RP2. The cognate G protein-GAP pair is conserved in yeast as Cin4-Cin2, and the ability of RP2 to act as a GAP can be correlated with its ability to complement a CIN2-deletion phenotype.
- Max-Planck-Institut für molekulare Physiologie, Abteilung Strukturelle Biologie, Otto-Hahn-Str. 11, 44227 Dortmund, Germany.
Organizational Affiliation: