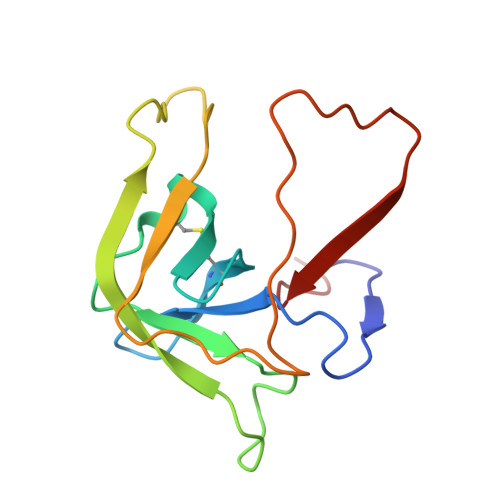
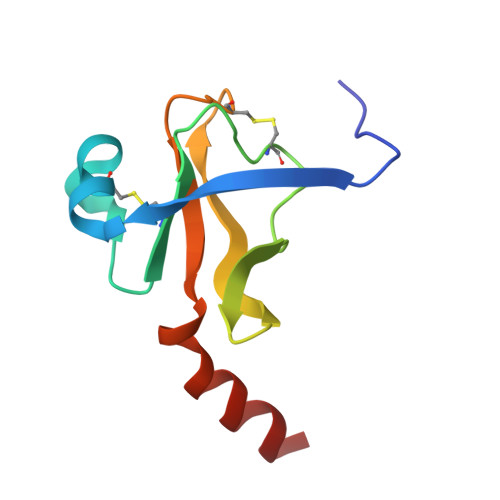
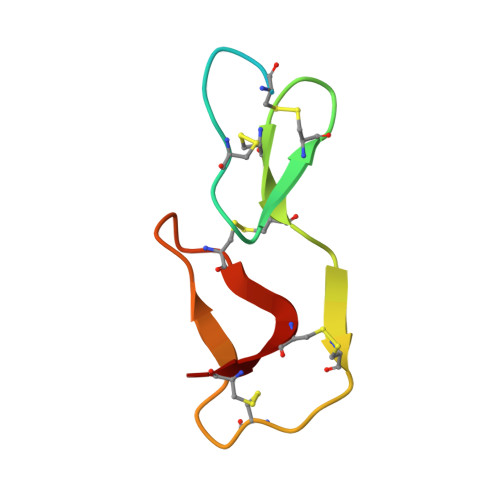
The crystal structure of guamerin in complex with chymotrypsin and the development of an elastase-specific inhibitor.
Kim, H., Chu, T.T.T., Kim, D.Y., Kim, D.R., Nguyen, C.M.T., Choi, J., Lee, J.R., Hahn, M.J., Kim, K.K.(2008) J Mol Biology 376: 184-192
- PubMed: 18155725
- DOI: https://doi.org/10.1016/j.jmb.2007.11.089
- Primary Citation of Related Structures:
3BG4 - PubMed Abstract:
Guamerin, a canonical serine protease inhibitor from Hirudo nipponia, was identified as an elastase-specific inhibitor and has potential application in various diseases caused by elevated elastase concentration. However, the application of guamerin is limited because it also shows inhibitory activity against other proteases. To improve the selectivity of guamerin as an elastase inhibitor, it is essential to understand the binding mode of the inhibitor to elastase and to other proteases. For this purpose, we determined the crystal structure of guamerin in complex with chymotrypsin at 2.5 A resolution. The binding mode of guamerin on elastase was explored from the model structure of guamerin/elastase. Guamerin binds to the hydrophobic pocket of the protease in a substrate-like manner using its binding loop. In order to improve the binding selectivity of guamerin to elastase, several residues in the binding loop were mutated and the inhibitory activities of the mutants against elastase and chymotrypsin were monitored. The substitution of the Met36 residue for Ala in the P1 site increased the inhibitory activity against elastase up to 14-fold, while the same mutant showed 7-fold decreased activity against chymotrypsin compared to the wild-type guamerin. Furthermore, the M36A guamerin mutant more effectively protected endothelial cells against cell damage caused by elastase than the wild-type guamerin.
- Department of Molecular Cell Biology, Samsung Biomedical Research Institute, Sungkyunkwan University School of Medicine, Suwon 440-746, Korea.
Organizational Affiliation: