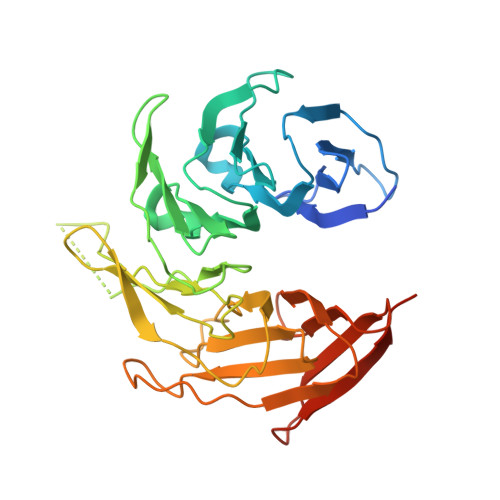
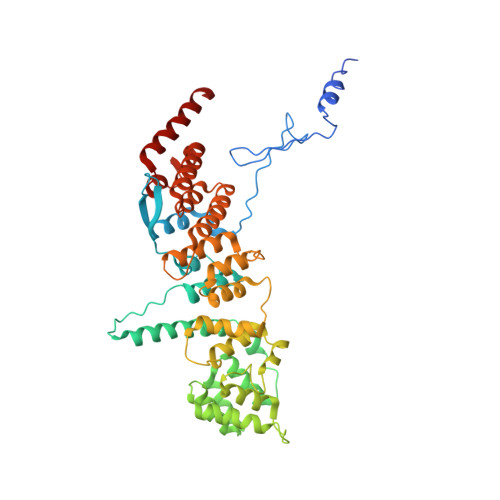
Architecture of a coat for the nuclear pore membrane.
Hsia, K.C., Stavropoulos, P., Blobel, G., Hoelz, A.(2007) Cell 131: 1313-1326
- PubMed: 18160040
- DOI: https://doi.org/10.1016/j.cell.2007.11.038
- Primary Citation of Related Structures:
3BG0, 3BG1 - PubMed Abstract:
The symmetric core of the nuclear pore complex can be considered schematically as a series of concentric cylinders. A peripheral cylinder coating the pore membrane contains the previously characterized, elongated heptamer that harbors Sec13-Nup145C in its middle section. Strikingly, Sec13-Nup145C crystallizes as a hetero-octamer in two space groups. Oligomerization of Sec13-Nup145C was confirmed biochemically. Importantly, the numerous interacting surfaces in the hetero-octamer are evolutionarily highly conserved, further underlining the physiological relevance of the oligomerization. The hetero-octamer forms a slightly curved, yet rigid rod of sufficient length to span the entire height of the proposed membrane-adjacent cylinder. In concordance with the dimensions and symmetry of the nuclear pore complex core, we suggest that the cylinder is constructed of four antiparallel rings, each ring being composed of eight heptamers arranged in a head-to-tail fashion. Our model proposes that the hetero-octamer would vertically traverse and connect the four stacked rings.
- Laboratory of Cell Biology, Howard Hughes Medical Institute, The Rockefeller University, 1230 York Avenue, New York, NY 10021, USA.
Organizational Affiliation: