The Heterodimeric Assembly of the CD94-NKG2 Receptor Family and Implications for Human Leukocyte Antigen-E Recognition
Sullivan, L.C., Clements, C.S., Beddoe, T., Johnson, D., Hoare, H.L., Lin, J., Huyton, T., Hopkins, E.J., Reid, H.H., Wilce, M.C.J., Kabat, J., Borrego, F., Coligan, J.E., Rossjohn, J., Brooks, A.G.(2007) Immunity 27: 900-911
- PubMed: 18083576
- DOI: https://doi.org/10.1016/j.immuni.2007.10.013
- Primary Citation of Related Structures:
3BDW - PubMed Abstract:
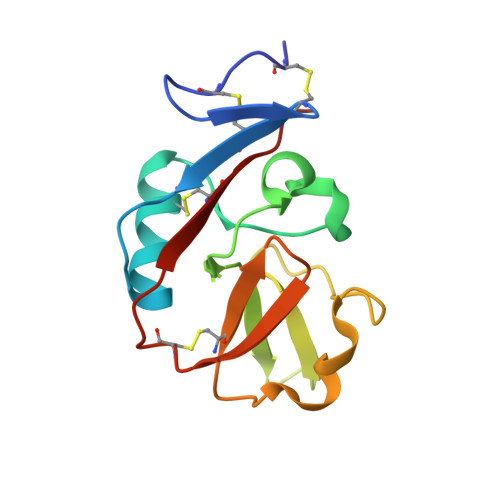
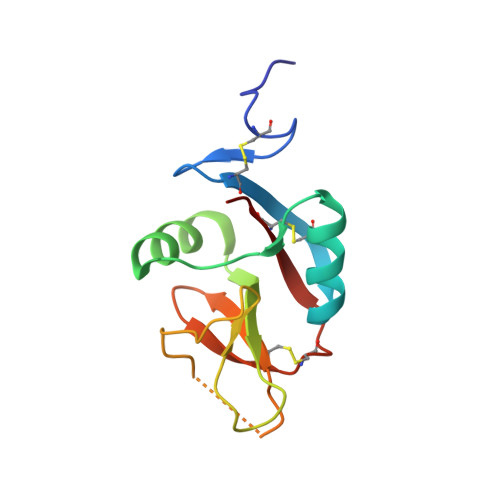
The CD94-NKG2 receptor family that regulates NK and T cells is unique among the lectin-like receptors encoded within the natural killer cell complex. The function of the CD94-NKG2 receptors is dictated by the pairing of the invariant CD94 polypeptide with specific NKG2 isoforms to form a family of functionally distinct heterodimeric receptors. However, the structural basis for this selective pairing and how they interact with their ligand, HLA-E, is unknown. We describe the 2.5 A resolution crystal structure of CD94-NKG2A in which the mode of dimerization contrasts with that of other homodimeric NK receptors. Despite structural homology between the CD94 and NKG2A subunits, the dimer interface is asymmetric, thereby providing a structural basis for the preferred heterodimeric assembly. Structure-based sequence comparisons of other CD94-NKG2 family members, combined with extensive mutagenesis studies on HLA-E and CD94-NKG2A, allows a model of the interaction between CD94-NKG2A and HLA-E to be established, in which the invariant CD94 chain plays a more dominant role in interacting with HLA-E in comparison to the variable NKG2 chain.
- Department of Microbiology and Immunology, University of Melbourne, Parkville, Victoria 3010, Australia.
Organizational Affiliation: