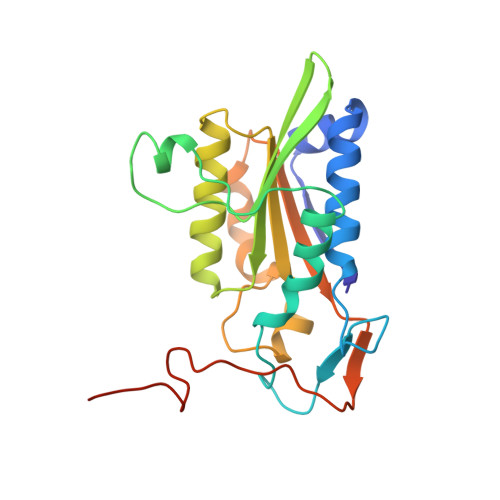
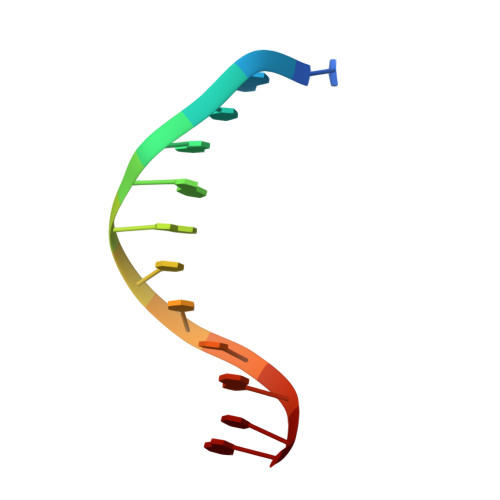
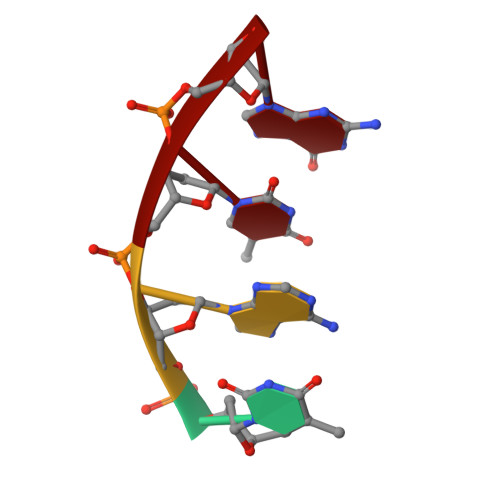
The role of metals in catalysis by the restriction endonuclease BamHI.
Viadiu, H., Aggarwal, A.K.(1998) Nat Struct Biol 5: 910-916
- PubMed: 9783752
- DOI: https://doi.org/10.1038/2352
- Primary Citation of Related Structures:
2BAM, 3BAM - PubMed Abstract:
Type II restriction enzymes are characterized by their remarkable specificity and simplicity. They require only divalent metals (such as Mg2+ or Mn2+) as cofactors to catalyze the hydrolysis of DNA. However, most of the structural work on endonucleases has been performed in the absence of metals, leaving unanswered questions about their mechanisms of DNA cleavage. Here we report structures of the endonuclease BamHI-DNA complex, determined in the presence of Mn2+ and Ca2+, that describe the enzyme at different stages of catalysis. Overall, the results support a two-metal mechanism of DNA cleavage for BamHI which is distinct from that of EcoRV.
- Department of Biochemistry and Molecular Biophysics, Columbia University, New York, New York 10032, USA.
Organizational Affiliation: