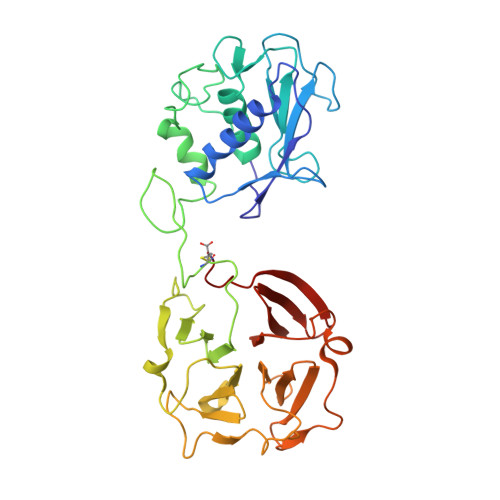
Evidence of reciprocal reorientation of the catalytic and hemopexin-like domains of full-length MMP-12.
Bertini, I., Calderone, V., Fragai, M., Jaiswal, R., Luchinat, C., Melikian, M., Mylonas, E., Svergun, D.I.(2008) J Am Chem Soc 130: 7011-7021
- PubMed: 18465858
- DOI: https://doi.org/10.1021/ja710491y
- Primary Citation of Related Structures:
2JXY, 3BA0 - PubMed Abstract:
The proteolytic activity of matrix metalloproteinases toward extracellular matrix components (ECM), cytokines, chemokines, and membrane receptors is crucial for several homeostatic and pathological processes. Active MMPs are a family of single-chain enzymes (23 family members in the human genome), most of which constituted by a catalytic domain and by a hemopexin-like domain connected by a linker. The X-ray structures of MMP-1 and MMP-2 suggest a conserved and well-defined spatial relationship between the two domains. Here we present structural data for MMP-12, suitably stabilized against self-hydrolysis, both in solution (NMR and SAXS) and in the solid state (X-ray), showing that the hemopexin-like and the catalytic domains experience conformational freedom with respect to each other on a time scale shorter than 10(-8) s. Hints on the probable conformations are also obtained. This experimental finding opens new perspectives for the often hypothesized active role of the hemopexin-like domain in the enzymatic activity of MMPs.
- Magnetic Resonance Center (CERM), University of Florence, Via L. Sacconi 6, 50019 Sesto Fiorentino, Italy. ivanobertini@cerm.unifi.it
Organizational Affiliation: