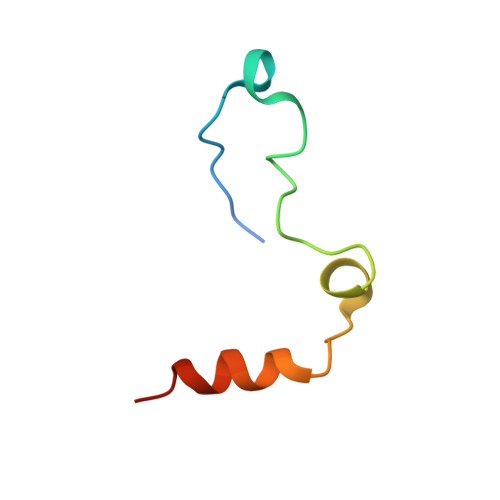
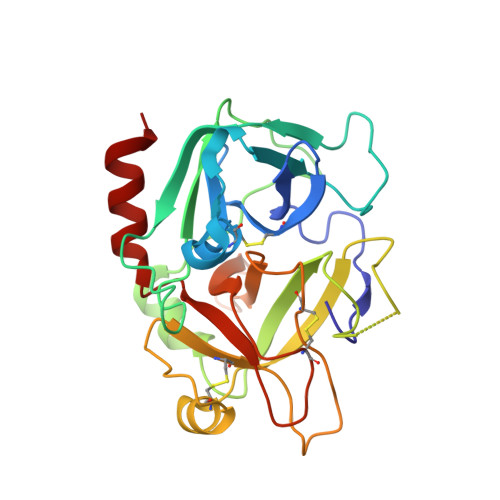
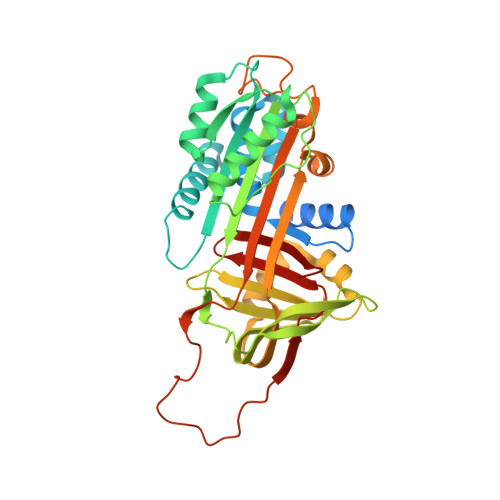
Molecular basis of thrombin recognition by protein C inhibitor revealed by the 1.6-A structure of the heparin-bridged complex.
Li, W., Adams, T.E., Nangalia, J., Esmon, C.T., Huntington, J.A.(2008) Proc Natl Acad Sci U S A 105: 4661-4666
- PubMed: 18362344
- DOI: https://doi.org/10.1073/pnas.0711055105
- Primary Citation of Related Structures:
3B9F - PubMed Abstract:
Protein C inhibitor (PCI) is a serpin with many roles in biology, including a dual role as pro- and anticoagulant in blood. The protease specificity and local function of PCI depend on its interaction with cofactors such as heparin-like glycosaminoglycans (GAGs) and thrombomodulin (TM). Both cofactors significantly increase the rate of thrombin inhibition, but GAGs serve to promote the anticoagulant activity of PCI, and TM promotes its procoagulant function. To gain insight into how PCI recognition of thrombin is aided by these cofactors, we determined a crystallographic structure of the Michaelis complex of PCI, thrombin, and heparin to 1.6 A resolution. Thrombin interacts with PCI in an unusual fashion that depends on the length of PCI's reactive center loop (RCL) to align the heparin-binding sites of the two proteins. The principal exosite contact is engendered by movement of thrombin's 60-loop in response to the unique P2 Phe of PCI. This mechanism of communication between the active site of thrombin and its recognition exosite is previously uncharacterized and may relate to other thrombin substrate-cofactor interactions. The cofactor activity of heparin thus depends on the formation of a heparin-bridged Michaelis complex and substrate-induced exosite contacts. We also investigated the cofactor effect of TM, establishing that TM bridges PCI to thrombin through additional direct interactions. A model of the PCI-thrombin-TM complex was built and evaluated by mutagenesis and suggests distinct binding sites for heparin and TM on PCI. These data significantly improve our understanding of the cofactor-dependent roles of PCI in hemostasis.
- Department of Haematology, Division of Structural Medicine, Thrombosis Research Unit, Cambridge Institute for Medical Research, University of Cambridge, Wellcome Trust/MRC Building, Hills Road, Cambridge CB2 0XY, United Kingdom.
Organizational Affiliation: