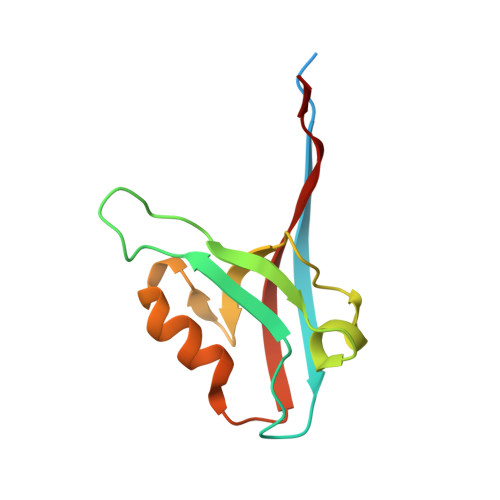
Crystal structure of the third PDZ domain of human ligand-of-numb protein-X (LNX1) in complex with the C-terminal peptide from the coxsackievirus and adenovirus receptor.
Ugochukwu, E., Burgess-Brown, N., Berridge, G., Elkins, J., Bunkoczi, G., Pike, A.C.W., Sundstrom, M., Arrowsmith, C.H., Weigelt, J., Edwards, A.M., Gileadi, O., von Delft, F., Doyle, D.To be published.