Pharmacology and in Vitro Profiling of a Novel Peroxisome Proliferator-Activated Receptor gamma Ligand, Cerco-A
Wakabayashi, K., Hayashi, S., Matsui, Y., Matsumoto, T., Furukawa, A., Kuroha, M., Tanaka, N., Inaba, T., Kanda, S., Tanaka, J., Okuyama, R., Wakimoto, S., Ogata, T., Araki, K., Ohsumi, J.(2011) Biol Pharm Bull 34: 1094-1104
- PubMed: 21720019
- DOI: https://doi.org/10.1248/bpb.34.1094
- Primary Citation of Related Structures:
3B1M - PubMed Abstract:
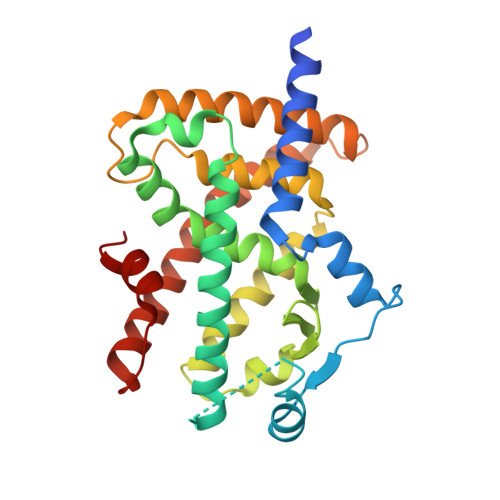
Peroxisome proliferator-activated receptor γ (PPARγ; NR1C3) is known as a key regulator of adipocytogenesis and the molecular target of thiazolidinediones (TZDs), also known as antidiabetic agents. Despite the clinical benefits of TZDs, their use is often associated with adverse effects including peripheral edema, congestive heart failure, and weight gain. Here we report the identification and characterization of a non-thiazolidinedione PPARγ partial agonist, Cerco-A, which is a derivative of the natural product, (-)-cercosporamide. Cerco-A was found to be a binder of the PPARγ ligand-binding domain in a ligand competitive binding assay and showed a unique cofactor recruitment profile compared to rosiglitazone. A crystal structure analysis revealed that Cerco-A binds to PPARγ without direct hydrogen bonding to helix12. In PPARγ transcriptional activation assay and an adipocyte differentiation assay, Cerco-A was a potent partial agonist of PPARγ. After a 14-day oral administration, once per day of Cerco-A in Zucker diabetic fatty (ZDF) rats, an apparent decrease of plasma glucose and triglyceride was observed, as with pioglitazone. To evaluate drug safety, Cerco-A was administered for 13 days orally in non-diabetic Zucker fatty (ZF) rats. Each of the hemodilution parameters (hematocrit, red blood cells number, and hemoglobin), which are considered as undesirable effects of TZDs, was improved significantly compared to pioglitazone. While Cerco-A showed body weight gain, as with pioglitazone, Cerco-A had significantly lower effects on heart and white adipose tissues weight gain. The results suggest that Cerco-A offers beneficial effects on glycemic control with attenuated undesirable side effects.
- Graduate School of Life and Environmental Sciences, University of Tsukuba, Tsukuba, Ibaraki 305–8577, Japan. wakabayashi.kenji.vy@rda.daiichisankyo.co.jp
Organizational Affiliation: