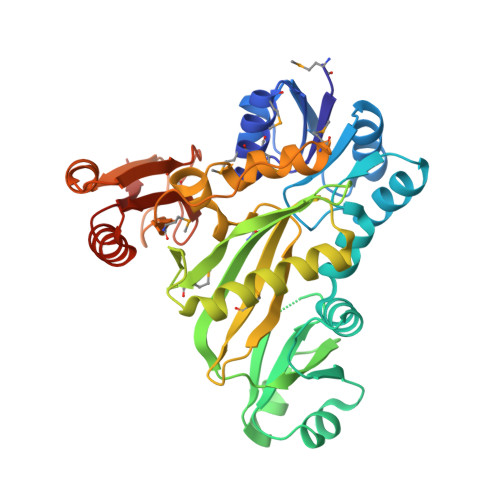
Crystal structures of N5-carboxyaminoimidazole ribonucleotide synthetase, PurK, from thermophilic bacteria
Okada, K., Taka, H., Tsunoda, S., Tamura, S., Miyazawa, R., Baba, S., Kanagawa, M., Nakagawa, N., Ebihara, A., Kuramitsu, S., Yokoyama, S., Kawai, G., Sampei, G.To be published.