Inverse agonist-like action of cadmium on G-protein-gated inward-rectifier K(+) channels
Inanobe, A., Matsuura, T., Nakagawa, A., Kurachi, Y.(2011) Biochem Biophys Res Commun 407: 366-371
- PubMed: 21396912
- DOI: https://doi.org/10.1016/j.bbrc.2011.03.025
- Primary Citation of Related Structures:
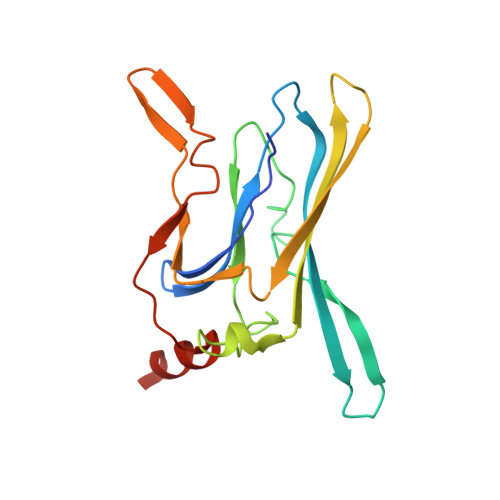
3AUW - PubMed Abstract:
The gate at the pore-forming domain of potassium channels is allosterically controlled by a stimulus-sensing domain. Using Cd²(+) as a probe, we examined the structural elements responsible for gating in an inward-rectifier K(+) channel (Kir3.2). One of four endogenous cysteines facing the cytoplasm contributes to a high-affinity site for inhibition by internal Cd²(+). Crystal structure of its cytoplasmic domain in complex with Cd²(+) reveals that octahedral coordination geometry supports the high-affinity binding. This mode of action causes the tethering of the N-terminus to CD loop in the stimulus-sensing domain, suggesting that their conformational changes participate in gating and Cd²(+) inhibits Kir3.2 by trapping the conformation in the closed state like "inverse agonist".
- Department of Pharmacology, Graduate School of Medicine, Osaka University, Osaka, Japan. inanobe@pharma2.med.osaka-u.ac.jp
Organizational Affiliation: