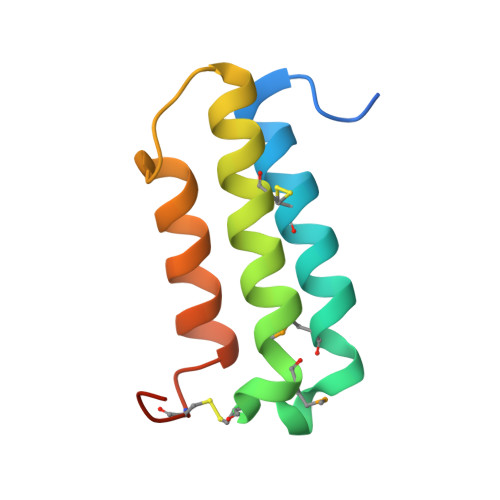
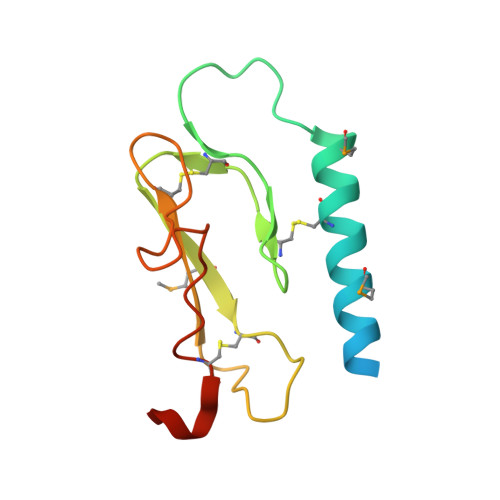
Structural basis for extracellular interactions between calcitonin receptor-like receptor and receptor activity-modifying protein 2 for adrenomedullin-specific binding
Kusano, S., Kukimoto-Niino, M., Hino, N., Ohsawa, N., Okuda, K., Sakamoto, K., Shirouzu, M., Shindo, T., Yokoyama, S.(2012) Protein Sci 21: 199-210
- PubMed: 22102369
- DOI: https://doi.org/10.1002/pro.2003
- Primary Citation of Related Structures:
3AQE, 3AQF - PubMed Abstract:
The calcitonin receptor-like receptor (CRLR), a class B GPCR, forms a heterodimer with receptor activity-modifying protein 2 (RAMP2), and serves as the adrenomedullin (AM) receptor to control neovascularization, while CRLR and RAMP1 form the calcitonin gene-related peptide (CGRP) receptor. Here, we report the crystal structures of the RAMP2 extracellular domain alone and in the complex with the CRLR extracellular domain. The CRLR-RAMP2 complex exhibits several intermolecular interactions that were not observed in the previously reported CRLR-RAMP1 complex, and thus the shape of the putative ligand-binding pocket of CRLR-RAMP2 is distinct from that of CRLR-RAMP1. The CRLR-RAMP2 interactions were confirmed for the full-length proteins on the cell surface by site-specific photo-crosslinking. Mutagenesis revealed that AM binding requires RAMP2 residues that are not conserved in RAMP1. Therefore, the differences in both the shapes and the key residues of the binding pocket are essential for the ligand specificity.
- RIKEN Systems and Structural Biology Center, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama, Japan.
Organizational Affiliation: