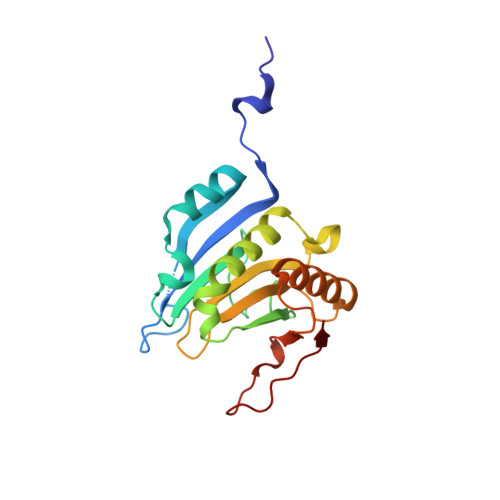
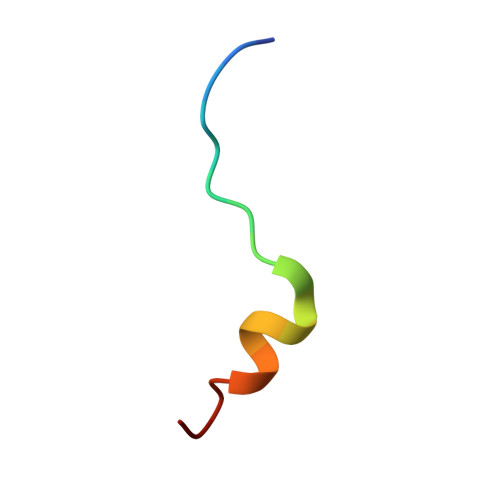
Structural scaffold for eIF4E binding selectivity of 4E-BP isoforms: crystal structure of eIF4E binding region of 4E-BP2 and its comparison with that of 4E-BP1.
Fukuyo, A., In, Y., Ishida, T., Tomoo, K.(2011) J Pept Sci 17: 650-657
- PubMed: 21661078
- DOI: https://doi.org/10.1002/psc.1384
- Primary Citation of Related Structures:
3AM7 - PubMed Abstract:
To clarify the higher eukaryotic initiation factor 4E (eIF4E) binding selectivity of 4E-binding protein 2 (4E-BP2) than of 4E-BP1, as determined by Trp fluorescence analysis, the crystal structure of the eIF4E binding region of 4E-BP2 in complex with m(7) GTP-bound human eIF4E has been determined by X-ray diffraction analysis and compared with that of 4E-BP1. The crystal structure revealed that the Pro47-Ser65 moiety of 4E-BP2 adopts a L-shaped conformation involving extended and α-helical structures and extends over the N-terminal loop and two different helix regions of eIF4E through hydrogen bonds, and electrostatic and hydrophobic interactions; these features were similarly observed for 4E-BP1. Although the pattern of the overall interaction of 4E-BP2 with eIF4E was similar to that of 4E-BP1, a notable difference was observed for the 60-63 sequence in relation to the conformation and binding selectivity of the 4E-BP isoform, i.e. Met-Glu-Cys-Arg for 4E-BP1 and Leu-Asp-Arg-Arg for 4E-BP2. In this paper, we report that the structural scaffold of the eIF4E binding preference for 4E-BP2 over 4E-BP1 is based on the stacking of the Arg63 planar side chain on the Trp73 indole ring of eIF4E and the construction of a compact hydrophobic space around the Trp73 indole ring by the Leu59-Leu60 sequence of 4E-BP2.
- Laboratory of Physical Chemistry, Osaka University of Pharmaceutical Sciences, Nasahara, Takatsuki, Japan.
Organizational Affiliation: