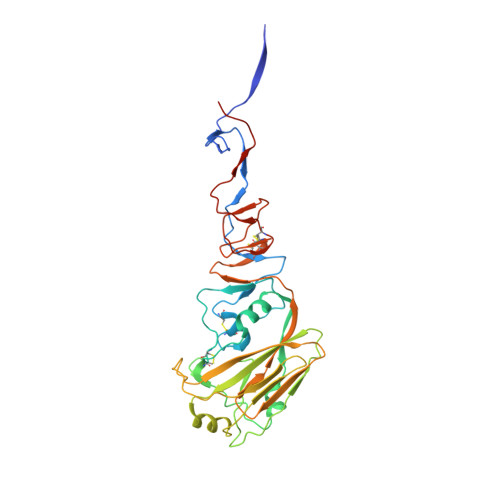
Crystal structure of the swine-origin A (H1N1)-2009 influenza A virus hemagglutinin (HA) reveals similar antigenicity to that of the 1918 pandemic virus
Zhang, W., Qi, J., Shi, Y., Li, Q., Gao, F., Sun, Y., Lu, X., Lu, Q., Vavricka, C.J., Liu, D., Yan, J., Gao, G.F.(2010) Protein Cell 1: 459-467
- PubMed: 21203961
- DOI: https://doi.org/10.1007/s13238-010-0059-1
- Primary Citation of Related Structures:
3AL4 - PubMed Abstract:
Influenza virus is the causative agent of the seasonal and occasional pandemic flu. The current H1N1 influenza pandemic, announced by the WHO in June 2009, is highly contagious and responsible for global economic losses and fatalities. Although the H1N1 gene segments have three origins in terms of host species, the virus has been named swine-origin influenza virus (S-OIV) due to a predominant swine origin. 2009 S-OIV has been shown to highly resemble the 1918 pandemic virus in many aspects. Hemagglutinin is responsible for the host range and receptor binding of the virus and is therefore a primary indicator for the potential of infection. Primary sequence analysis of the 2009 S-OIV hemagglutinin (HA) reveals its closest relationship to that of the 1918 pandemic influenza virus, however, analysis at the structural level is necessary to critically assess the functional significance. In this report, we report the crystal structure of soluble hemagglutinin H1 (09H1) at 2.9 Å, illustrating that the 09H1 is very similar to the 1918 pandemic HA (18H1) in overall structure and the structural modules, including the five defined antiboby (Ab)-binding epitopes. Our results provide an explanation as to why sera from the survivors of the 1918 pandemics can neutralize the 2009 S-OIV, and people born around the 1918 are resistant to the current pandemic, yet younger generations are more susceptible to the 2009 pandemic.
- CAS Key Laboratory of Pathogenic Microbiology and Immunology, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, China.
Organizational Affiliation: