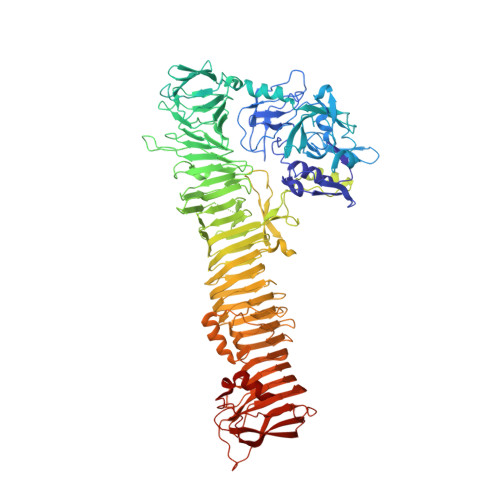
Role of domains within the autotransporter Hbp/Tsh
Nishimura, K., Yoon, Y.-H., Kurihara, A., Unzai, S., Luirink, J., Park, S.-Y., Tame, J.R.H.(2010) Acta Crystallogr D Biol Crystallogr 66: 1295-1300
- PubMed: 21123869
- DOI: https://doi.org/10.1107/S0907444910036966
- Primary Citation of Related Structures:
3AK5 - PubMed Abstract:
The autotransporter Tsh (temperature-sensitive haemagglutinin) secreted by avian pathogenic Escherichia coli was reported in 1994 and the almost identical Hbp (haemoglobin protease) was discovered some years later in isolates from patients suffering from peritoneal abscesses. However, the function of the protein remains uncertain. The crystal structure of Hbp shows that the protein carries a serine protease domain (domain 1) and a small domain of 75 residues called domain 2 which is inserted into the long β-helix characteristic of autotransporter passenger proteins. In this paper, domain 1 is shown to bind calcium, although metal ions binding to this site do not seem to regulate protease activity. Tsh has been reported to bind red cells and components of the extracellular matrix, but it is demonstrated that these properties are not a consequence of the presence of domain 2.
- Yokohama City University, Suehiro 1-7-29, Tsurumi, Yokohama 230-0045, Japan.
Organizational Affiliation: