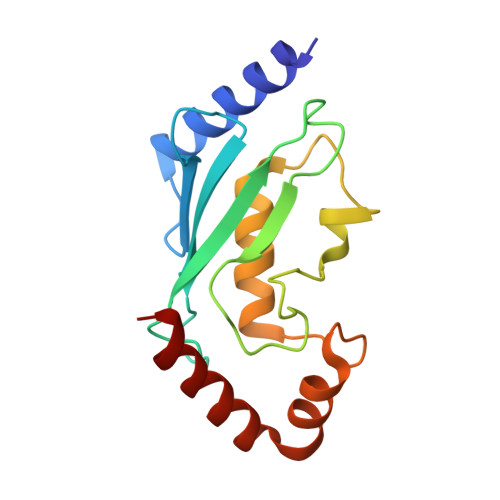
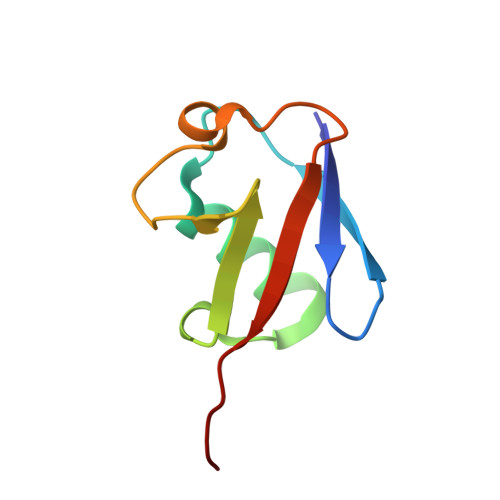
Crystal Structure of UbcH5b~Ubiquitin Intermediate: Insight into the Formation of the Self-Assembled E2~Ub Conjugates
Sakata, E., Satoh, T., Yamamoto, S., Yamaguchi, Y., Yagi-Utsumi, M., Kurimoto, E., Tanaka, K., Wakatsuki, S., Kato, K.(2010) Structure 18: 138-147
- PubMed: 20152160
- DOI: https://doi.org/10.1016/j.str.2009.11.007
- Primary Citation of Related Structures:
3A33 - PubMed Abstract:
E2 ubiquitin-conjugating enzymes catalyze the attachment of ubiquitin to lysine residues of target proteins. The UbcH5b E2 enzyme has been shown to play a key role in the initiation of the ubiquitination of substrate proteins upon action of several E3 ligases. Here we have determined the 2.2 A crystal structure of an intermediate of UbcH5b~ubiquitin (Ub) conjugate, which is assembled into an infinite spiral through the backside interaction. This active complex may provide multiple E2 active sites, enabling efficient ubiquitination of substrates. Indeed, biochemical assays support a model in which the self-assembled UbcH5b~Ub can serve as a bridge for the gap between the lysine residue of the substrate and the catalytic cysteine of E2.
- Department of Structural Biology and Biomolecular Engineering, Graduate School of Pharmaceutical Sciences, Nagoya City University, 3-1 Tanabe-dori, Mizuho-ku, Nagoya 467-8603, Japan.
Organizational Affiliation: