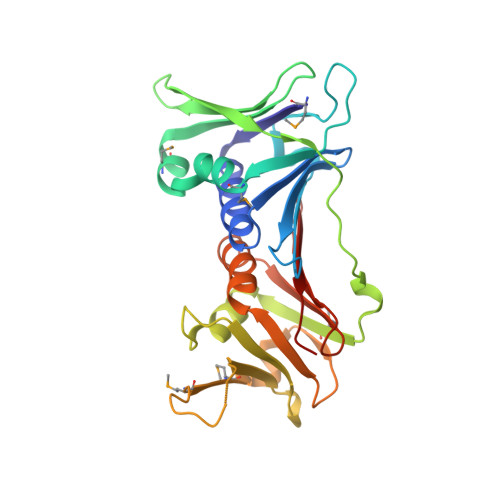
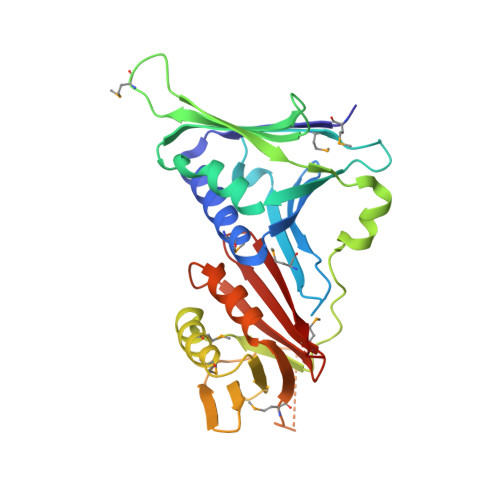
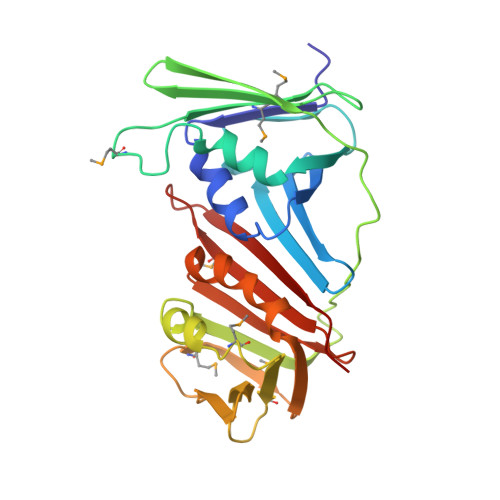
Crystal Structure of the Human Rad9-Hus1-Rad1 Clamp
Sohn, S.Y., Cho, Y.(2009) J Mol Biology 390: 490-502
- PubMed: 19464297
- DOI: https://doi.org/10.1016/j.jmb.2009.05.028
- Primary Citation of Related Structures:
3A1J - PubMed Abstract:
Three evolutionarily conserved proteins, Rad9, Hus1, and Rad1, form a heterotrimeric 9-1-1 complex that plays critical roles in cellular responses to DNA damage by activating checkpoints and by recruiting DNA repair enzymes to DNA lesions. We have determined the crystal structure of the human Rad9 (residues 1-272)-Hus1-Rad1 complex at 2.5 A resolution. The 9(1-272)-1-1 complex forms a closed ring, with each subunit having a similar structure. Despite its high level of similarity to proliferating cell nucleus antigen in terms of overall structure, the 9(1-272)-1-1 complex exhibits notable differences in local structures, including interdomain connecting loops, H2 and H3 helices, and loops in the vicinity of the helices of each subunit. These local structural variations provide several unique features to the 9-1-1 heterotrimeric complex-including structures of intermolecular interfaces and the inner surface around the central hole, and different electrostatic potentials at and near the interdomain connecting loops of each 9-1-1 subunit-compared to the proliferating cell nucleus antigen trimer. We propose that these structural features allow the 9-1-1 complex to bind to a damaged DNA during checkpoint control and to serve as a platform for base excision repair. We also show that the 9(1-272)-1-1 complex, but not the full-length 9-1-1 complex, forms a stable complex with the 5' recessed DNA, suggesting that the C-terminal tail of Rad9 is involved in the regulation of the 9-1-1 complex in DNA binding.
- National Creative Research Center for Structural Biology and Department of Life Science, Pohang University of Science and Technology, Hyo-ja dong, Pohang, KyungBook, South Korea.
Organizational Affiliation: