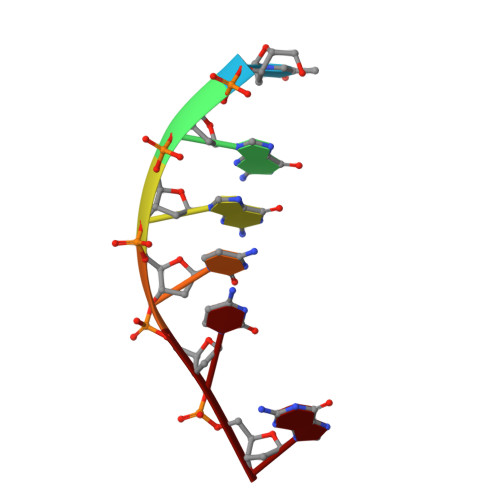
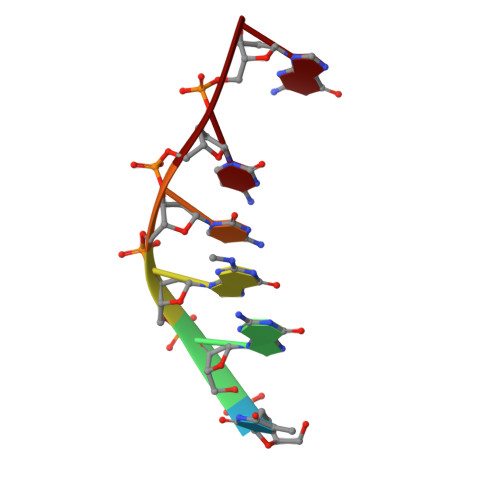
Binding of the modified daunorubicin WP401 adjacent to a T-G base pair induces the reverse Watson-Crick conformation: crystal structures of the WP401-TGGCCG and WP401-CGG[br5C]CG complexes.
Dutta, R., Gao, Y.G., Priebe, W., Wang, A.H.(1998) Nucleic Acids Res 26: 3001-3005
- PubMed: 9611247
- DOI: https://doi.org/10.1093/nar/26.12.3001
- Primary Citation of Related Structures:
380D, 381D - PubMed Abstract:
2'-Bromo-4'-epi-daunorubicin (alpha-manno configuration, denoted WP401) is a new anthracycline drug that exhibits promising activity toward multidrug-resistant cancer cells. We carried out X-ray diffraction analyses of the complexes formed in the presence of formaldehyde between WP401 and two DNA hexamers, TGGCCG and CGG[br5C]CG. The two complexes crystallized in different crystal lattices with respective crystal data of space group P4322, a = b = 37.20 A, c = 70.53 A and space group P43212, a = b = 37.23 A, c = 61. 96 A. These new crystal forms are different from the P41212 form of other daunorubicin/doxorubicin complexes studied previously. The refined crystal structures at approximately 2.0 A resolution revealed that the entire 2:1 drug-DNA complex is in the asymmetrical unit. Two WP401 drug molecules bind to the duplex, with the aglycones intercalated between the CpG or TpG steps and their modified daunosamines in the minor groove. As observed earlier, in the presence of formaldehyde, WP401 more readily forms a covalent adduct with (C/T)GG*:CCG than with (C/T)GC:G*CG (G* is the crosslink site), the opposite of what is seen for daunorubicin and doxorubicin. Surprisingly, the two T-G mismatched base pairs in the WP401-TGGCCG complex adopt the reverse Watson-Crick conformation, instead of the wobble conformation. The unusual T-G reverse Watson-Crick conformation may be required in order to maintain favorable stacking interactions between the base pair and the aglycone of WP401. Our results show that chemical modifications like bromo or iodo substitution on anthracycline drugs have significant effects on their DNA binding properties.
- Department of Cell and Structural Biology, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA.
Organizational Affiliation: