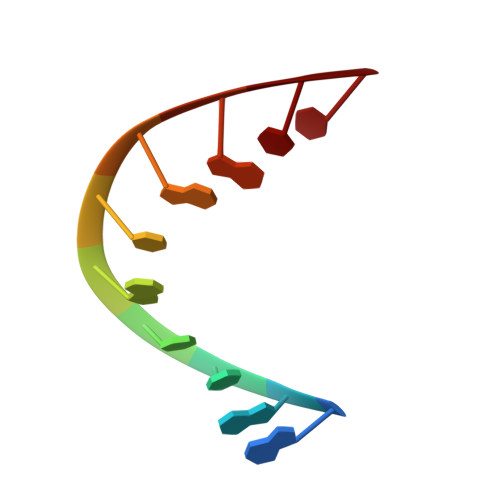
X-ray crystal structures of half the human papilloma virus E2 binding site: d(GACCGCGGTC).
Finley, J.B., Luo, M.(1998) Nucleic Acids Res 26: 5719-5727
- PubMed: 9838004
- DOI: https://doi.org/10.1093/nar/26.24.5719
- Primary Citation of Related Structures:
348D, 349D, 401D - PubMed Abstract:
The X-ray crystal structure of the DNA decamer d(GACCGCGGTC), containing half the human papilloma virus E2 binding site, has been solved from two crystals grown at different ionic conditions (50 mM MgCl2and 50 mM spermine or 1.56 mM MgCl2and 1.56 mM spermine). Despite the variation in salt concentration, the two DNA structures are in a very similar, A-type DNA conformation, with helical axes curving towards the major groove. Although the salt concentrations do not effect the helical parameters or hydration to a large degree, there is a change in the overall helical curvature; 18 degrees and 31 degrees for the low and high salt structures, respectively. This curvature appears to be sequence specific and biologically relevant when compared with similar DNA structures, including the E2 binding site of a protein-DNA complex.
- Center for Macromolecular Crystallography, Department of Microbiology, University of Alabama-Birmingham, Birmingham, AL 35294-0005, USA. jfinley@orion.cmc.uab.edu
Organizational Affiliation: