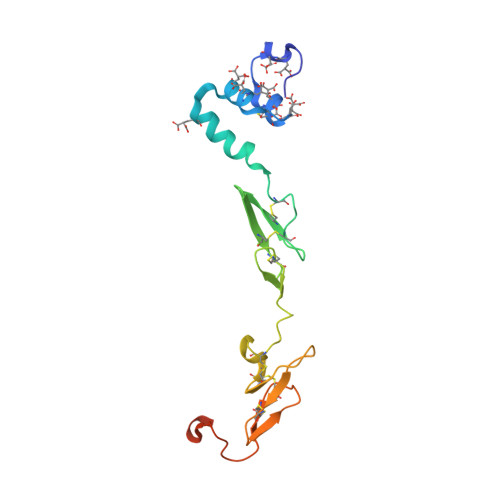
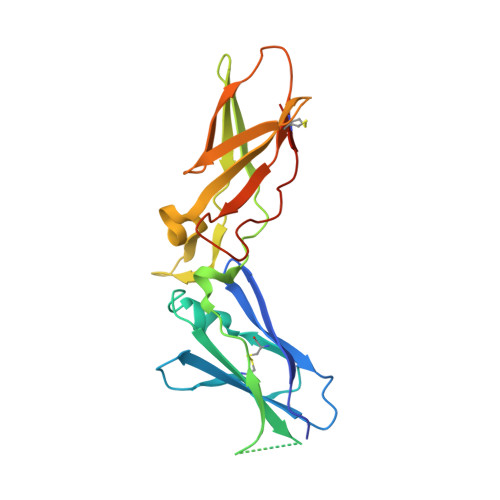
Peptide Mimetic Factor VIIa Inhibitor: Importance of Hydrophilic Pocket in S2 Site to Improve Selectivity aganist Thrombin
Kadono, S., Sakamoto, A., Kikuchi, Y., Oh-eda, M., Yabuta, N., Koga, T., Hattori, K., Shiraishi, T., Haramura, M., Kodama, H., Esaki, T., Sato, H., Watanabe, S., Itoh, S., Ohta, M., Kozono, T.(2005) Lett Drug Des Discov 2: 177-181