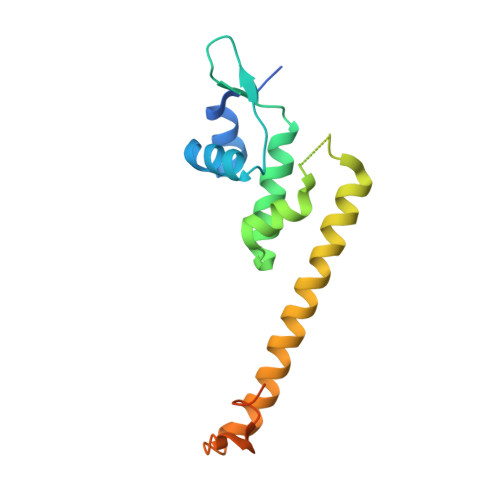
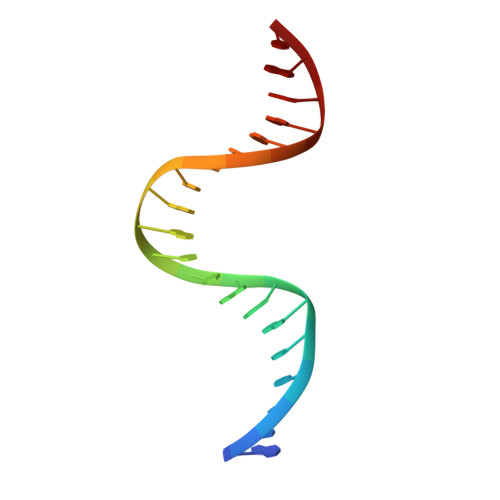
Crystal structure of the [2Fe-2S] oxidative-stress sensor SoxR bound to DNA
Watanabe, S., Kita, A., Kobayashi, K., Miki, K.(2008) Proc Natl Acad Sci U S A 105: 4121-4126
- PubMed: 18334645
- DOI: https://doi.org/10.1073/pnas.0709188105
- Primary Citation of Related Structures:
2ZHG, 2ZHH - PubMed Abstract:
The [2Fe-2S] transcription factor SoxR, a member of the MerR family, functions as a bacterial sensor of oxidative stress such as superoxide and nitric oxide. SoxR is activated by reversible one-electron oxidation of the [2Fe-2S] cluster and then enhances the production of various antioxidant proteins through the soxRS regulon. In the active state, SoxR and other MerR family proteins activate transcription from unique promoters, which have a long 19- or 20-bp spacer between the -35 and -10 operator elements, by untwisting the promoter DNA. Here, we show the crystal structures of SoxR and its complex with the target promoter in the oxidized (active) state. The structures reveal that the [2Fe-2S] cluster of SoxR is completely solvent-exposed and surrounded by an asymmetric environment stabilized by interaction with the other subunit. The asymmetrically charged environment of the [2Fe-2S] cluster probably causes redox-dependent conformational changes of SoxR and the target promoter. Compared with the promoter structures with the 19-bp spacer previously studied, the DNA structure is more sharply bent, by approximately 1 bp, with the two central base pairs holding Watson-Crick base pairs. Comparison of the target promoter sequences of the MerR family indicates that the present DNA structure represents the activated conformation of the target promoter with a 20-bp spacer in the MerR family.
- Department of Chemistry, Graduate School of Science, Kyoto University, Sakyo-ku, Kyoto 606-8502, Japan.
Organizational Affiliation: