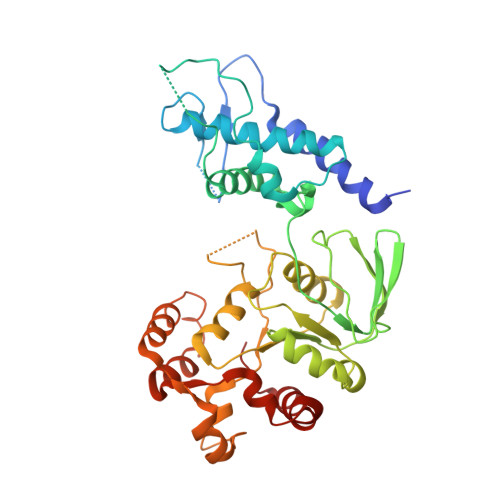
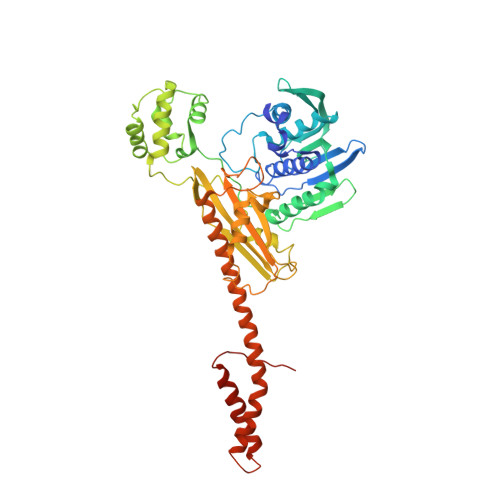
Crystal Structure of an Intact Type II DNA Topoisomerase: Insights into DNA Transfer Mechanisms
Graille, M., Durand, D., Lecointe, F., Gadelle, D., Quevillon-Cheruel, S., Vachette, P., Forterre, P., van Tilbeurgh, H.(2008) Structure 16: 360-370
- PubMed: 18334211
- DOI: https://doi.org/10.1016/j.str.2007.12.020
- Primary Citation of Related Structures:
2ZBK - PubMed Abstract:
DNA topoisomerases resolve DNA topological problems created during transcription, replication, and recombination. These ubiquitous enzymes are essential for cell viability and are highly potent targets for the development of antibacterial and antitumoral drugs. Type II enzymes catalyze the transfer of a DNA duplex through another one in an ATP-dependent mechanism. Because of its small size and sensitivity to antitumoral drugs, the archaeal DNA topoisomerase VI, a type II enzyme, is an excellent model for gaining further understanding of the organization and mechanism of these enzymes. We present the crystal structure of intact DNA topoisomerase VI bound to radicicol, an inhibitor of human topo II, and compare it to the conformation of the apo-protein as determined by small-angle X-ray scattering in solution. This structure, combined with a wealth of experimental data gathered on these enzymes, allows us to propose a structural model for the two-gate DNA transfer mechanism.
- Institut de Biochimie et de Biophysique Moléculaire et Cellulaire, UMR8619 CNRS, Université Paris-Sud, IFR115, F-91405 Orsay, France.
Organizational Affiliation: