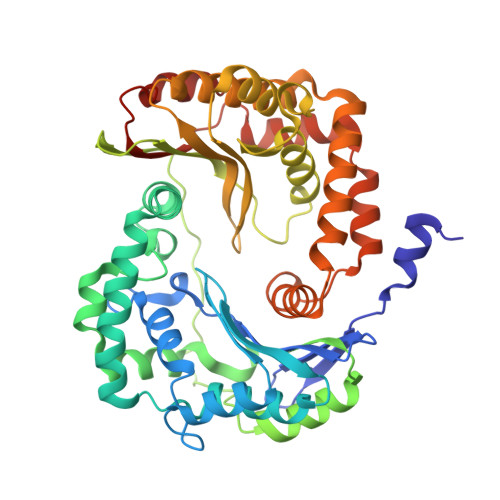
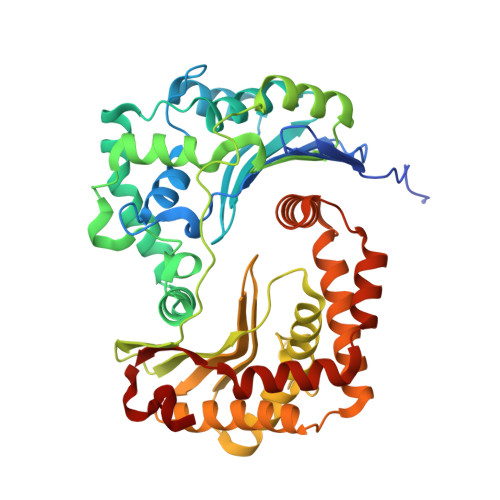
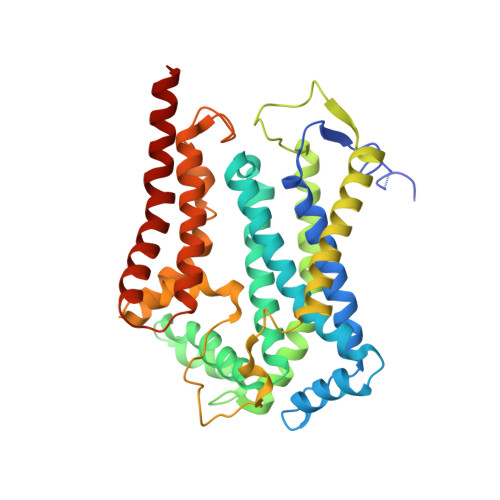
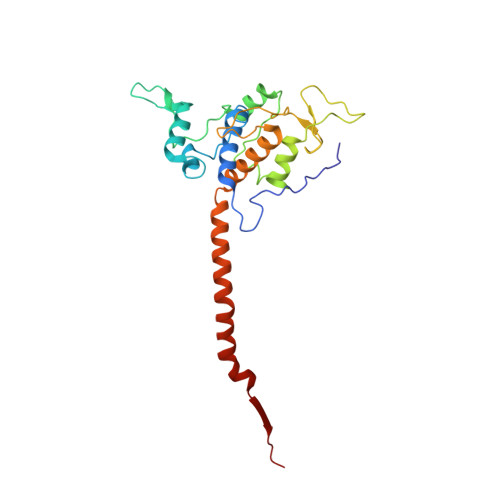
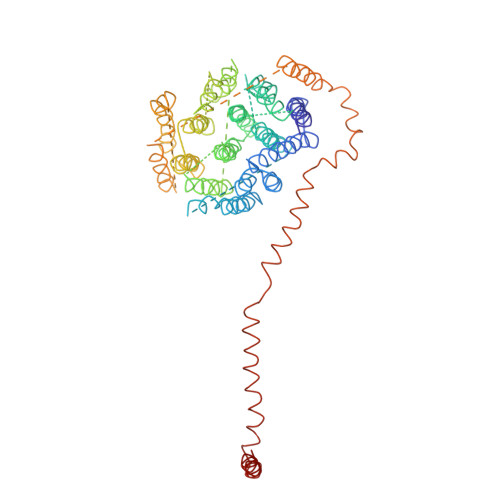
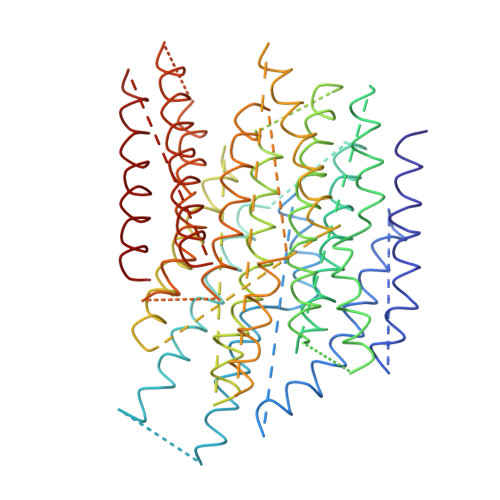
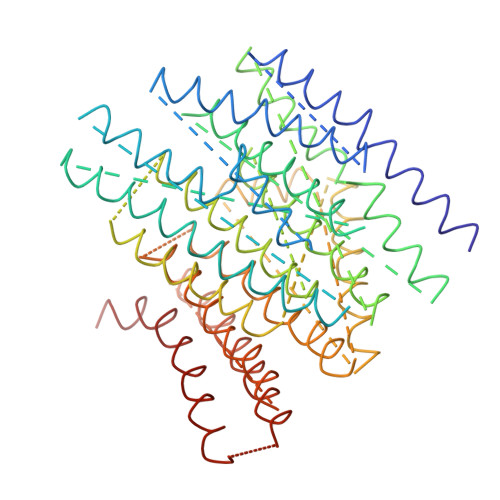
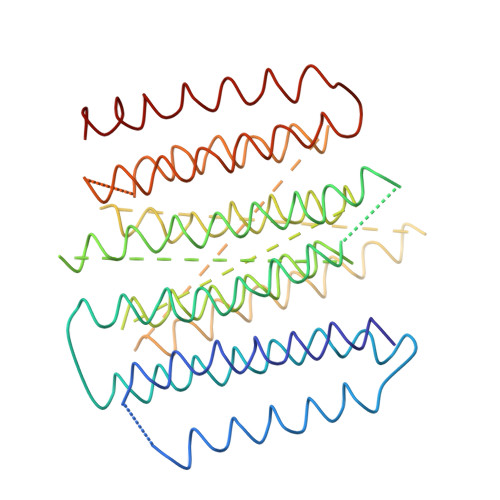
Arrangement of Electron Transport Chain Components in Bovine Mitochondrial Supercomplex I(1)III(2)Iv(1).
Althoff, T., Mills, D.J., Popot, J.-L., Kuehlbrandt, W.(2011) EMBO J 30: 4652
- PubMed: 21909073
- DOI: https://doi.org/10.1038/emboj.2011.324
- Primary Citation of Related Structures:
2YBB - PubMed Abstract:
The respiratory chain in the inner mitochondrial membrane contains three large multi-enzyme complexes that together establish the proton gradient for ATP synthesis, and assemble into a supercomplex. A 19-Å 3D map of the 1.7-MDa amphipol-solubilized supercomplex I(1)III(2)IV(1) from bovine heart obtained by single-particle electron cryo-microscopy reveals an amphipol belt replacing the membrane lipid bilayer. A precise fit of the X-ray structures of complex I, the complex III dimer, and monomeric complex IV indicates distances of 13 nm between the ubiquinol-binding sites of complexes I and III, and of 10-11 nm between the cytochrome c binding sites of complexes III and IV. The arrangement of respiratory chain complexes suggests two possible pathways for efficient electron transfer through the supercomplex, of which the shorter branch through the complex III monomer proximal to complex I may be preferred.
- Abteilung Strukturbiologie, Max-Planck-Institut für Biophysik, Frankfurt, Germany.
Organizational Affiliation: