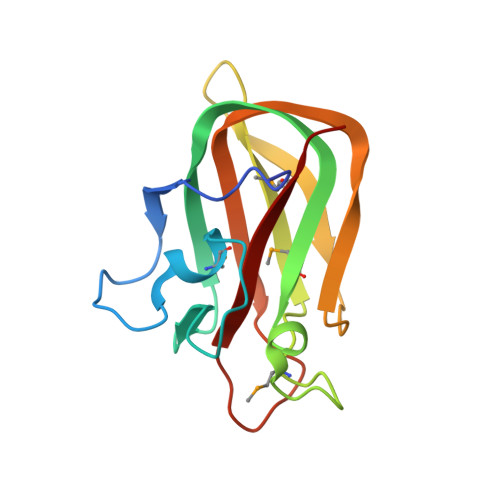
Structural Analysis of a Putative Family 32 Carbohydrate-Binding Module from the Streptococcus Pneumoniae Enzyme Endod.
Abbott, D.W., Boraston, A.B.(2011) Acta Crystallogr Sect F Struct Biol Cryst Commun 67: 429
- PubMed: 21505233
- DOI: https://doi.org/10.1107/S1744309111001874
- Primary Citation of Related Structures:
2XQX - PubMed Abstract:
EndoD is an architecturally complex endo-β-1,4-N-acetylglucosamidase from Streptococcus pneumoniae that cleaves the chitobiose core of N-linked glycans and contributes to pneumococcal virulence. Although the glycoside hydrolase family 85 catalytic module has been structurally and functionally characterized, nothing is known about the ancillary modules and how they contribute to the overall function of the enzyme. Presented here is the 2.0 Å resolution structure of a family 32 carbohydrate-binding module of EndoD, SpCBM32, solved by single-wavelength anomalous dispersion. The putative binding site of this protein is a charge-neutral relatively flat region on the protein surface that contains one prominently exposed tryptophan residue that extends into the solvent. These topographical features are discussed in the biological context of EndoD activity and a hypothesis is made about the complex structure of its potential carbohydrate ligand.
- Department of Biochemistry and Microbiology, University of Victoria, PO Box 3055 STN CSC, Victoria, BC V8W 3P6, Canada.
Organizational Affiliation: