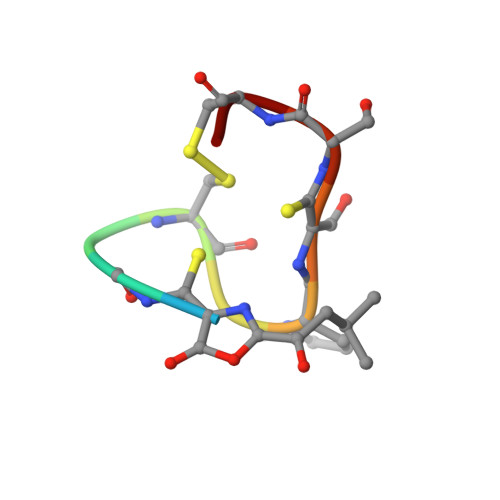
Copper-Binding Properties and Structures of Methanobactins from Methylosinus Trichosporium Ob3B.
El Ghazouani, A., Basle, A., Firbank, S.J., Knapp, C.W., Gray, J., Graham, D.W., Dennison, C.(2011) Inorg Chem 50: 1378
- PubMed: 21254756
- DOI: https://doi.org/10.1021/ic101965j
- Primary Citation of Related Structures:
2XJH, 2XJI - PubMed Abstract:
Methanobactins (mbs) are a class of copper-binding peptides produced by aerobic methane oxidizing bacteria (methanotrophs) that have been linked to the substantial copper needs of these environmentally important microorganisms. The only characterized mbs are those from Methylosinus trichosporium OB3b and Methylocystis strain SB2. M. trichosporium OB3b produces a second mb (mb-Met), which is missing the C-terminal Met residue from the full-length form (FL-mb). The as-isolated copper-loaded mbs bind Cu(I). The absence of the Met has little influence on the structure of the Cu(I) site, and both molecules mediate switchover from the soluble iron methane mono-oxygenase to the particulate copper-containing enzyme in M. trichosporium OB3b cells. Cu(II) is reduced in the presence of the mbs under our experimental conditions, and the disulfide plays no role in this process. The Cu(I) affinities of these molecules are extremely high with values of (6-7) × 10(20) M(-1) determined at pH ≥ 8.0. The affinity for Cu(I) is 1 order of magnitude lower at pH 6.0. The reduction potentials of copper-loaded FL-mb and mb-Met are 640 and 590 mV respectively, highlighting the strong preference for Cu(I) and indicating different Cu(II) affinities for the two forms. Cleavage of the disulfide bridge results in a decrease in the Cu(I) affinity to ∼9 × 10(18) M(-1) at pH 7.5. The two thiolates can also bind Cu(I), albeit with much lower affinity (∼ 3 × 10(15) M(-1) at pH 7.5). The high affinity of mbs for Cu(I) is consistent with a physiological role in copper uptake and protection.
- Institute for Cell and Molecular Biosciences, Medical School, Newcastle University, Newcastle upon Tyne NE2 4HH, United Kingdom.
Organizational Affiliation: