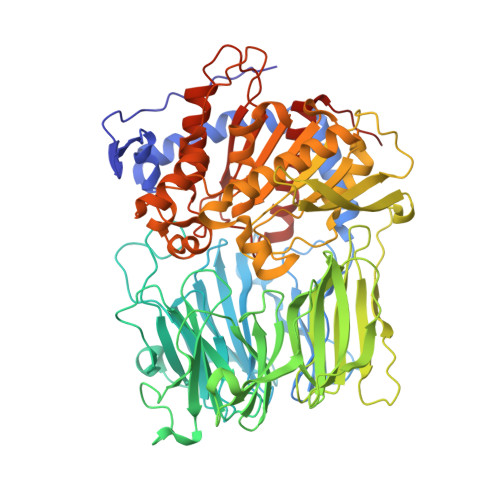
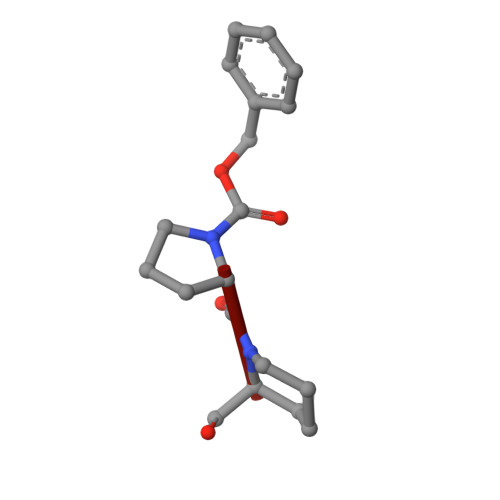
Inhibition of Prolyl Oligopeptidase with a Synthetic Unnatural Dipeptide
Racys, D.T., Rea, D., Fulop, V., Wills, M.(2010) Bioorg Med Chem 18: 4775
- PubMed: 20627594
- DOI: https://doi.org/10.1016/j.bmc.2010.05.012
- Primary Citation of Related Structures:
2XDW - PubMed Abstract:
A new inhibitor, containing a linked proline-piperidine structure, for the enzyme prolyl oligopeptidase (POP) has been synthesised and demonstrated to bind covalently with the enzyme at the active site. This provides evidence that covalent inhibitors of POP do not have to be limited to structures containing five-membered N-containing heterocyclic rings.
- Department of Chemistry, The University of Warwick, Coventry CV47AL, UK.
Organizational Affiliation: