Domain Cross-Talk During Effector Binding to the Multidrug Binding Ttgr Regulator.
Daniels, C., Daddaoua, A., Lu, D., Zhang, X., Ramos, J.L.(2010) J Biological Chem 285: 21372
- PubMed: 20435893
- DOI: https://doi.org/10.1074/jbc.M110.113282
- Primary Citation of Related Structures:
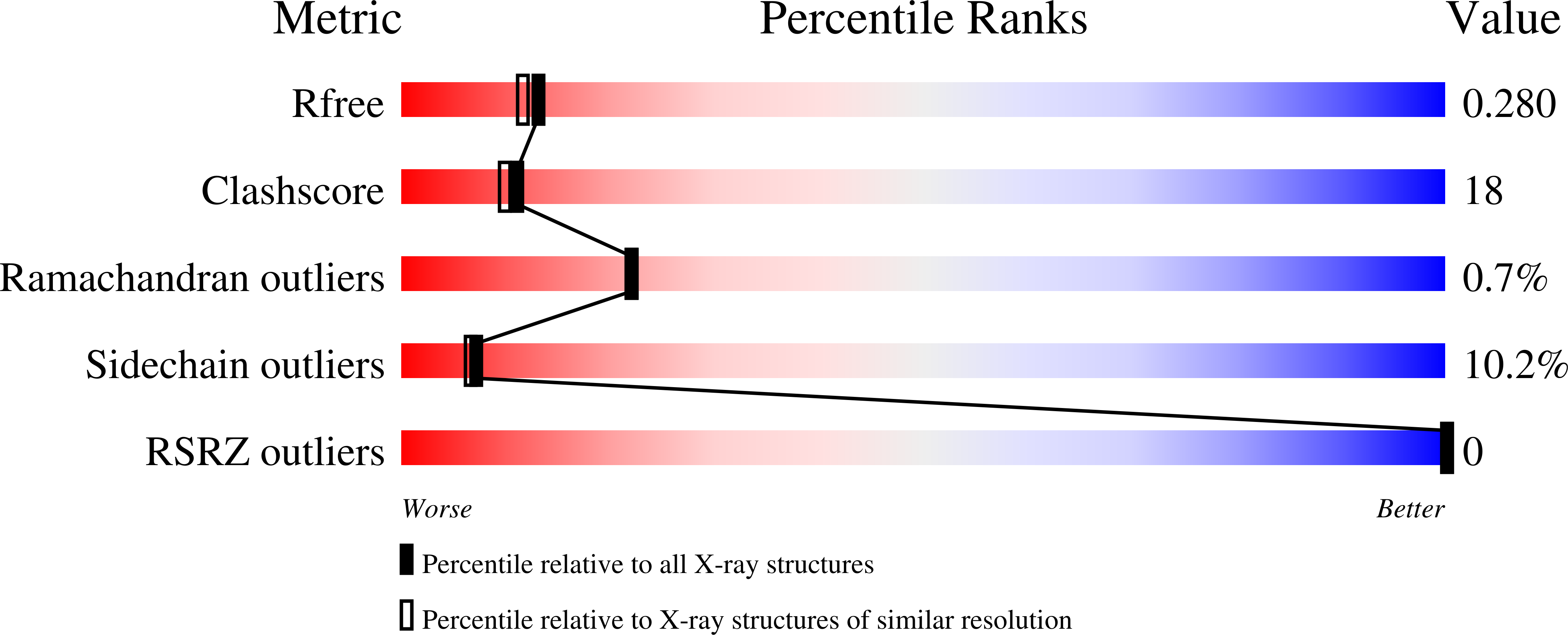
2XDN - PubMed Abstract:
A major mechanism of antibiotic resistance in bacteria is the active extrusion of toxic compounds through membrane-bound efflux pumps. The TtgR protein represses transcription of ttgABC, a key efflux pump in Pseudomonas putida DOT-T1E capable of extruding antibiotics, solvents, and flavonoids. TtgR contains two distinct and overlapping ligand binding sites, one is broad and contains mainly hydrophobic residues, whereas the second is deep and contains polar residues. Mutants in the ligand binding pockets were generated and characterized using electrophoretic mobility shift assays, isothermal titration calorimetry, and promoter expression. Several mutants were affected in their response to effectors in vitro: mutants H70A, H72A, and R75A did not dissociate from promoter DNA in the presence of chloramphenicol. Other mutants exhibited altered binding to the operator: L66A and L66AV96A mutants bound 3- and 15-fold better than the native protein, whereas the H67A mutant bound with 3-fold lower affinity. In vivo expression assays using a fusion of the promoter of ttgA to lacZ and antibiotic tolerance correlated with the in vitro observations, namely that mutant H67A leads to increased basal expression levels and enhances antibiotic tolerance, whereas mutants L66A and L66AV96A exhibit lower basal expression levels and decreased resistance to antibiotics. The crystal structure of TtgR H67A was resolved. The data provide evidence for the inter-domain communication that is predicted to be required for the transmission of the effector binding signal to the DNA binding domain and provide important information to understand TtgR/DNA/effector interactions.
- Department of Environmental Protection, CSIC, E-18008 Granada, Spain.
Organizational Affiliation: