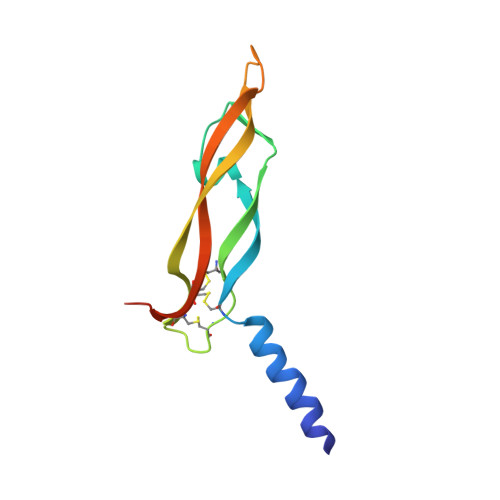
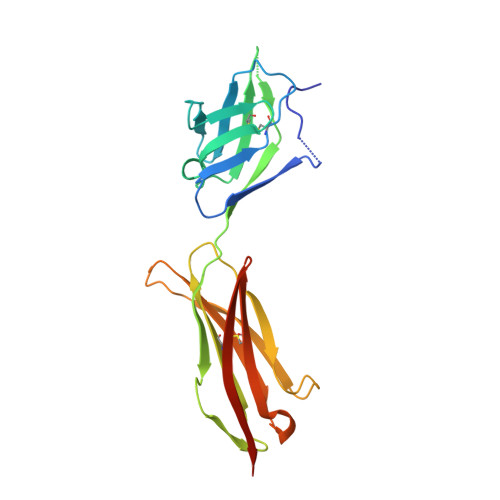
Structural Determinants of Growth Factor Binding and Specificity by Vegf Receptor 2.
Leppanen, V.-M., Prota, A.E., Jeltsch, M., Anisimov, A., Kalkkinen, N., Strandin, T., Lankinen, H., Goldman, A., Ballmer-Hofer, K., Alitalo, K.(2010) Proc Natl Acad Sci U S A 107: 2425
- PubMed: 20145116
- DOI: https://doi.org/10.1073/pnas.0914318107
- Primary Citation of Related Structures:
2X1W, 2X1X - PubMed Abstract:
Vascular endothelial growth factors (VEGFs) regulate blood and lymph vessel formation through activation of three receptor tyrosine kinases, VEGFR-1, -2, and -3. The extracellular domain of VEGF receptors consists of seven immunoglobulin homology domains, which, upon ligand binding, promote receptor dimerization. Dimerization initiates transmembrane signaling, which activates the intracellular tyrosine kinase domain of the receptor. VEGF-C stimulates lymphangiogenesis and contributes to pathological angiogenesis via VEGFR-3. However, proteolytically processed VEGF-C also stimulates VEGFR-2, the predominant transducer of signals required for physiological and pathological angiogenesis. Here we present the crystal structure of VEGF-C bound to the VEGFR-2 high-affinity-binding site, which consists of immunoglobulin homology domains D2 and D3. This structure reveals a symmetrical 22 complex, in which left-handed twisted receptor domains wrap around the 2-fold axis of VEGF-C. In the VEGFs, receptor specificity is determined by an N-terminal alpha helix and three peptide loops. Our structure shows that two of these loops in VEGF-C bind to VEGFR-2 subdomains D2 and D3, while one interacts primarily with D3. Additionally, the N-terminal helix of VEGF-C interacts with D2, and the groove separating the two VEGF-C monomers binds to the D2/D3 linker. VEGF-C, unlike VEGF-A, does not bind VEGFR-1. We therefore created VEGFR-1/VEGFR-2 chimeric proteins to further study receptor specificity. This biochemical analysis, together with our structural data, defined VEGFR-2 residues critical for the binding of VEGF-A and VEGF-C. Our results provide significant insights into the structural features that determine the high affinity and specificity of VEGF/VEGFR interactions.
- Molecular Cancer Biology Program, Biomedicum Helsinki, Department of Pathology, Haartman Institute and Helsinki University Central Hospital, PO Box 63, University of Helsinki, Haartmaninkatu 8, FI-00014 Helsinki, Finland.
Organizational Affiliation: