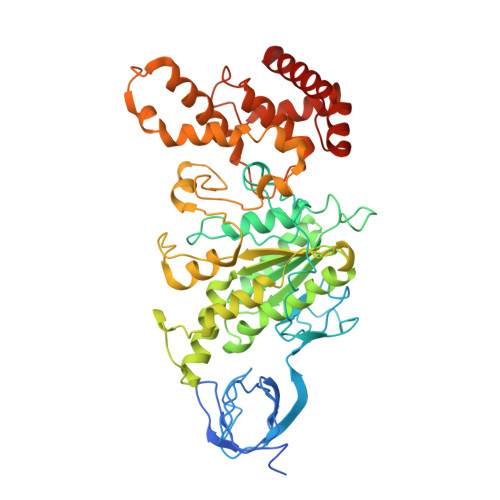
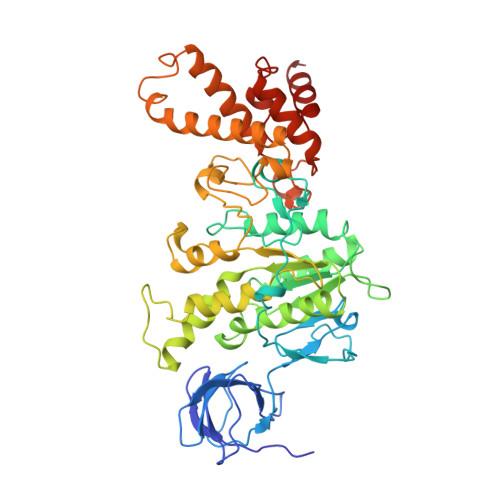
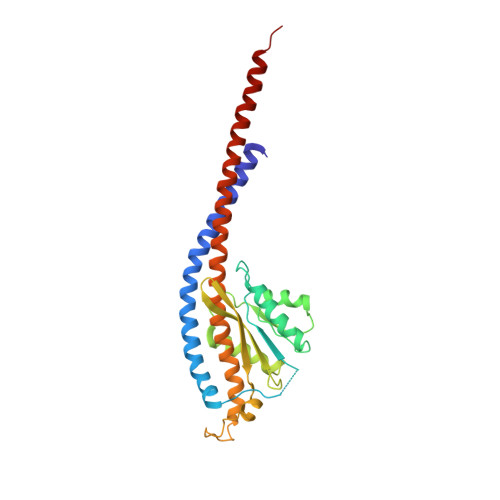
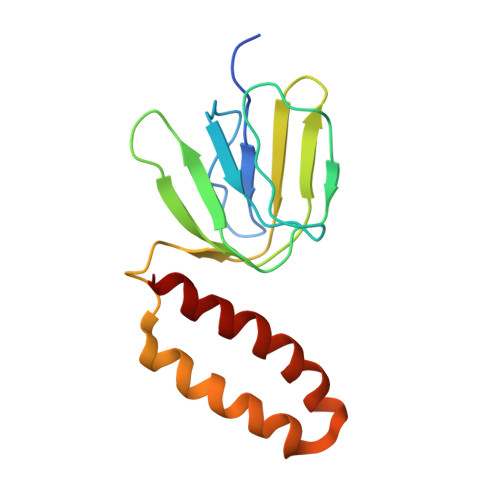
Crystal Structure of the Mg.Adp-Inhibited State of the Yeast F1C10-ATP Synthase.
Dautant, A., Velours, J., Giraud, M.(2010) J Biological Chem 285: 29502
- PubMed: 20610387
- DOI: https://doi.org/10.1074/jbc.M110.124529
- Primary Citation of Related Structures:
2WPD - PubMed Abstract:
The F(1)c(10) subcomplex of the yeast F(1)F(0)-ATP synthase includes the membrane rotor part c(10)-ring linked to a catalytic head, (αβ)(3), by a central stalk, γδε. The Saccharomyces cerevisiae yF(1)c(10)·ADP subcomplex was crystallized in the presence of Mg·ADP, dicyclohexylcarbodiimide (DCCD), and azide. The structure was solved by molecular replacement using a high resolution model of the yeast F(1) and a bacterial c-ring model with 10 copies of the c-subunit. The structure refined to 3.43-Å resolution displays new features compared with the original yF(1)c(10) and with the yF(1) inhibited by adenylyl imidodiphosphate (AMP-PNP) (yF(1)(I-III)). An ADP molecule was bound in both β(DP) and β(TP) catalytic sites. The α(DP)-β(DP) pair is slightly open and resembles the novel conformation identified in yF(1), whereas the α(TP)-β(TP) pair is very closed and resembles more a DP pair. yF(1)c(10)·ADP provides a model of a new Mg·ADP-inhibited state of the yeast F(1). As for the original yF(1) and yF(1)c(10) structures, the foot of the central stalk is rotated by ∼40 ° with respect to bovine structures. The assembly of the F(1) central stalk with the F(0) c-ring rotor is mainly provided by electrostatic interactions. On the rotor ring, the essential cGlu(59) carboxylate group is surrounded by hydrophobic residues and is not involved in hydrogen bonding.
- Université Bordeaux 2, CNRS, Institut de Biochimie et Génétique Cellulaires, 1 rue Camille Saint-Saëns, 33077 Bordeaux Cedex, France. A.Dautant@ibgc.cnrs.fr
Organizational Affiliation: