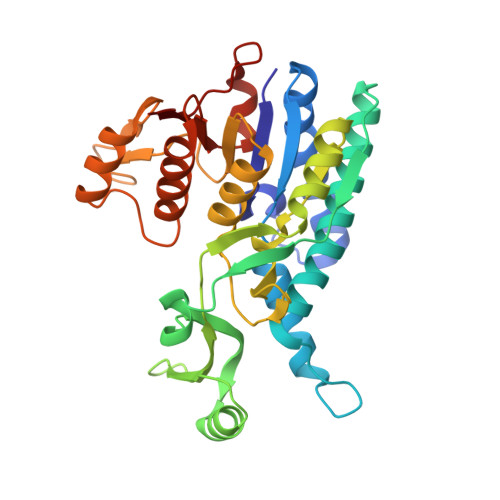
Substrate Binding and Catalysis in Carbamate Kinase Ascertained by Crystallographic and Site- Directed Mutagenesis Studies. Movements and Significance of a Unique Globular Subdomain of This Key Enzyme for Fermentative ATP Production in Bacteria.
Ramon-Maiques, S., Marina, A., Guinot, A., Gil-Ortiz, F., Uriarte, M., Fita, I., Rubio, V.(2010) J Mol Biology 397: 1261
- PubMed: 20188742
- DOI: https://doi.org/10.1016/j.jmb.2010.02.038
- Primary Citation of Related Structures:
2WE4, 2WE5 - PubMed Abstract:
Carbamate kinase (CK) makes ATP from ADP and carbamoyl phosphate (CP) in the final step of the microbial fermentative catabolism of arginine, agmatine, and oxalurate/allantoin. Two previously reported CK structures failed to clarify CP binding and catalysis and to reveal the significance of the protruding subdomain (PSD) that hangs over the CK active center as an exclusive and characteristic CK feature. We clarify now these three questions by determining two crystal structures of Enterococcus faecalis CK (one at 1.5 A resolution and containing bound MgADP, and the other at 2.1 A resolution and having in the active center one sulfate and two fixed water molecules that mimic one bound CP molecule) and by mutating active-center residues, determining the consequences of these mutations on enzyme functionality. Superimposition of the present crystal structures reconstructs the filled active center in the ternary complex, immediately suggesting in-line associative phosphoryl group transfer and a mechanism for enzyme catalysis involving N51, K209, K271, D210, and the PSD residue K128. The large respective increases and decreases in K(m)(CP) and k(cat) triggered by the mutations N51A, K128A, K209A, and D210N corroborate the ternary complex active-site architecture and the catalytic mechanism proposed. The extreme negative effects of K128A demonstrate a key role of the PSD in substrate binding and catalysis. The crystal structures reveal large rigid-body movements of the PSD towards the enzyme body that place K128 next to CP and bury the CP site. A mechanism that connects CP site occupation with the PSD approach, involving V206-I207 in the CP site and P162-S163 in the PSD stem, is identified. The effects of the V206A and V206L mutations support this mechanism. It is concluded that the PSD movement allows CK to select against the abundant CP/carbamate analogues acetylphosphate/acetate and bicarbonate, rendering CK highly selective for CP/carbamate.
- Instituto de Biomedicina de Valencia-Consejo Superior de Investigaciones Científicas and Centro de Investigación Biomédica en Red de Enfermedades Raras (Instituto de Salud Carlos III), Jaime Roig 11, Valencia 46010, Spain.
Organizational Affiliation: