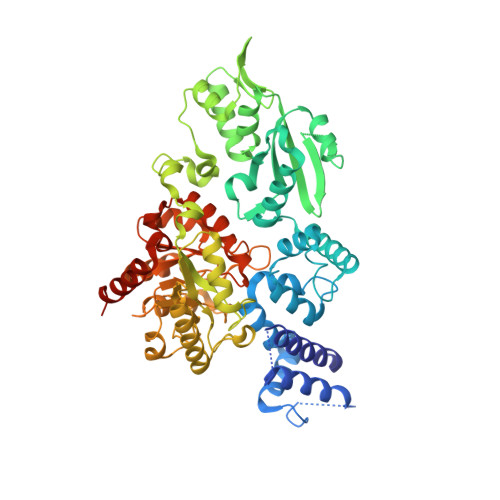
The Family X DNA Polymerase from Deinococcus Radioduran Adopts a Non-Standard Extended Conformation.
Leulliot, N., Cladiere, L., Lecointe, F., Durand, D., Hubscher, U., Van Tilbeurgh, H.(2009) J Biological Chem 284: 11992
- PubMed: 19251692
- DOI: https://doi.org/10.1074/jbc.M809342200
- Primary Citation of Related Structures:
2W9M - PubMed Abstract:
Deinococcus radiodurans is an extraordinarily radioresistant bacterium that is able to repair hundreds of radiation-induced double-stranded DNA breaks. One of the players in this pathway is an X family DNA polymerase (PolX(Dr)). Deletion of PolX(Dr) has been shown to decrease the rate of repair of double-stranded DNA breaks and increase cell sensitivity to gamma-rays. A 3'-->5' exonuclease activity that stops cutting close to DNA loops has also been demonstrated. The present crystal structure of PolX(Dr) solved at 2.46-A resolution reveals that PolX(Dr) has a novel extended conformation in stark contrast to the closed "right hand" conformation commonly observed for DNA polymerases. This extended conformation is stabilized by the C-terminal PHP domain, whose putative nuclease active site is obstructed by its interaction with the polymerase domain. The overall conformation and the presence of non standard residues in the active site of the polymerase X domain makes PolX(Dr) the founding member of a novel class of polymerases involved in DNA repair but whose detailed mode of action still remains enigmatic.
- Institut de Biochimie et de Biophysique Moléculaire et Cellulaire, Université de Paris-Sud, CNRS, UMR8619, IFR115, Bât 430, Orsay 91405 Cedex, France.
Organizational Affiliation: