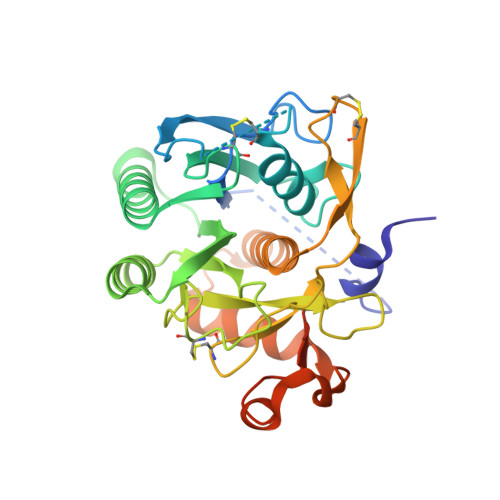
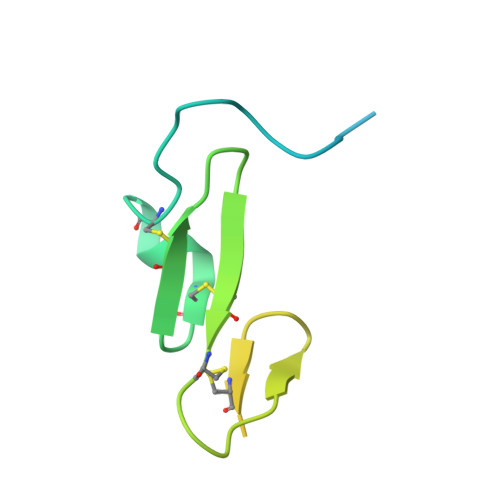
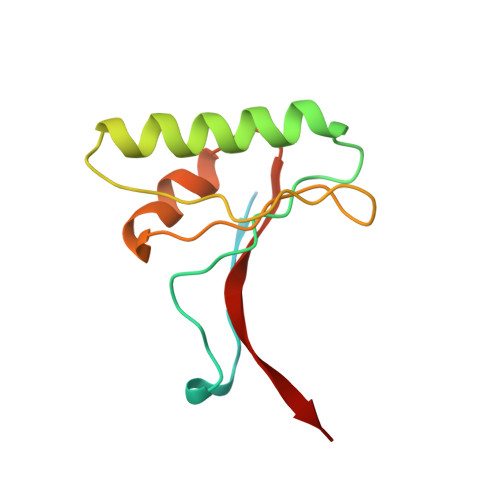
Structural and Biochemical Characterization of the Wild Type Pcsk9/Egf-Ab Complex and Natural Fh Mutants.
Bottomley, M.J., Cirillo, A., Orsatti, L., Ruggeri, L., Fisher, T.S., Santoro, J.C., Cummings, R.T., Cubbon, R.M., Lo Surdo, P., Calzetta, A., Noto, A., Baysarowich, J., Mattu, M., Talamo, F., De Francesco, R., Sparrow, C.P., Sitlani, A., Carfi, A.(2009) J Biological Chem 284: 1313
- PubMed: 19001363
- DOI: https://doi.org/10.1074/jbc.M808363200
- Primary Citation of Related Structures:
2W2M, 2W2N, 2W2O, 2W2P, 2W2Q - PubMed Abstract:
PCSK9 regulates low density lipoprotein receptor (LDLR) levels and consequently is a target for the prevention of atherosclerosis and coronary heart disease. Here we studied the interaction, of LDLR EGF(A/AB) repeats with PCSK9. We show that PCSK9 binds the EGF(AB) repeats in a pH-dependent manner. Although the PCSK9 C-terminal domain is not involved in LDLR binding, PCSK9 autocleavage is required. Moreover, we report the x-ray structure of the PCSK9DeltaC-EGF(AB) complex at neutral pH. Compared with the low pH PCSK9-EGF(A) structure, the new structure revealed rearrangement of the EGF(A) His-306 side chain and disruption of the salt bridge with PCSK9 Asp-374, thus suggesting the basis for enhanced interaction at low pH. In addition, the structure of PCSK9DeltaC bound to EGF(AB)(H306Y), a mutant associated with familial hypercholesterolemia (FH), reveals that the Tyr-306 side chain forms a hydrogen bond with PCSK9 Asp-374, thus mimicking His-306 in the low pH conformation. Consistently, Tyr-306 confers increased affinity for PCSK9. Importantly, we found that although the EGF(AB)(H306Y)-PCSK9 interaction is pH-independent, LDLR(H306Y) binds PCSK9 50-fold better at low pH, suggesting that factors other than His-306 contribute to the pH dependence of PCSK9-LDLR binding. Further, we determined the structures of EGF(AB) bound to PCSK9DeltaC containing the FH-associated D374Y and D374H mutations, revealing additional interactions with EGF(A) mediated by Tyr-374/His-374 and providing a rationale for their disease phenotypes. Finally, we report the inhibitory properties of EGF repeats in a cellular assay measuring LDL uptake.
- Department of Biochemistry, Istituto di Ricerca di Biologia Molecolare "P. Angeletti", Via Pontina Km 30.600, 00040 Pomezia (Rome), Italy. matthew_bottomley@merck.com
Organizational Affiliation: