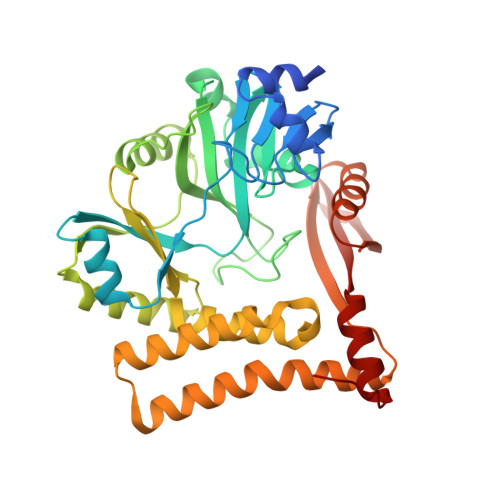
The structure of an archaeal homodimeric ligase which has RNA circularization activity.
Brooks, M.A., Meslet-Cladiere, L., Graille, M., Kuhn, J., Blondeau, K., Myllykallio, H., van Tilbeurgh, H.(2008) Protein Sci 17: 1336-1345
- PubMed: 18511537
- DOI: https://doi.org/10.1110/ps.035493.108
- Primary Citation of Related Structures:
2VUG - PubMed Abstract:
The genome of Pyrococcus abyssi contains two open reading frames encoding proteins which had been previously predicted to be DNA ligases, Pab2002 and Pab1020. We show that while the former is indeed a DNA ligase, Pab1020 had no effect on the substrate deoxyoligo-ribonucleotides tested. Instead, Pab1020 catalyzes the nucleotidylation of oligo-ribonucleotides in an ATP-dependent reaction, suggesting that it is an RNA ligase. We have solved the structure of Pab1020 in complex with the ATP analog AMPPNP by single-wavelength anomalous dispersion (SAD), elucidating a structure with high structural similarity to the catalytic domains of two RNA ligases from the bacteriophage T4. Additional carboxy-terminal domains are also present, and one of these mediates contacts with a second protomer, which is related by noncrystallographic symmetry, generating a homodimeric structure. These C-terminal domains are terminated by short domain swaps which themselves end within 5 A of the active sites of the partner molecules. Additionally, we show that the protein is indeed capable of circularizing RNA molecules in an ATP-dependent reaction. These structural and biochemical results provide an insight into the potential physiological roles of Pab1020.
- IBBMC-CNRS, Université de Paris-Sud, CNRS-UMR8619, IFR115, 91405 Orsay, France.
Organizational Affiliation: