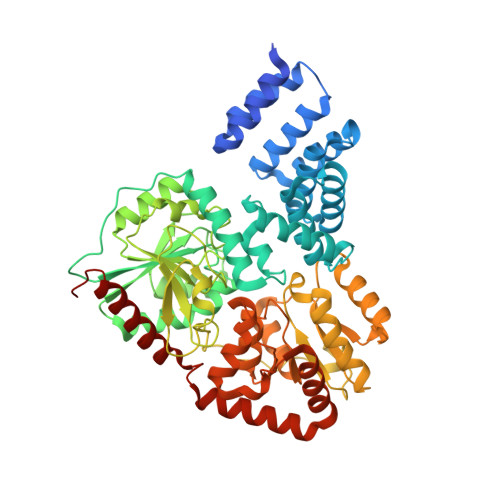
Structural Insights Into Mechanism and Specificity of O-Glcnac Transferase.
Clarke, A.J., Hurtado-Guerrero, R., Pathak, S., Schuttelkopf, A.W., Borodkin, V., Shepherd, S.M., Ibrahim, A.F.M., Van Aalten, D.M.F.(2008) EMBO J 27: 2780
- PubMed: 18818698
- DOI: https://doi.org/10.1038/emboj.2008.186
- Primary Citation of Related Structures:
2JLB, 2VSY - PubMed Abstract:
Post-translational modification of protein serines/threonines with N-acetylglucosamine (O-GlcNAc) is dynamic, inducible and abundant, regulating many cellular processes by interfering with protein phosphorylation. O-GlcNAcylation is regulated by O-GlcNAc transferase (OGT) and O-GlcNAcase, both encoded by single, essential, genes in metazoan genomes. It is not understood how OGT recognises its sugar nucleotide donor and performs O-GlcNAc transfer onto proteins/peptides, and how the enzyme recognises specific cellular protein substrates. Here, we show, by X-ray crystallography and mutagenesis, that OGT adopts the (metal-independent) GT-B fold and binds a UDP-GlcNAc analogue at the bottom of a highly conserved putative peptide-binding groove, covered by a mobile loop. Strikingly, the tetratricopeptide repeats (TPRs) tightly interact with the active site to form a continuous 120 A putative interaction surface, whereas the previously predicted phosphatidylinositide-binding site locates to the opposite end of the catalytic domain. On the basis of the structure, we identify truncation/point mutants of the TPRs that have differential effects on activity towards proteins/peptides, giving first insights into how OGT may recognise its substrates.
- Division of Biological Chemistry & Drug Discovery, College of Life Sciences, University of Dundee, Dundee, UK.
Organizational Affiliation: