Modified Uridines with C5-Methylene Substituents at the First Position of the tRNA Anticodon Stabilize U.G Wobble Pairing During Decoding.
Kurata, S., Weixlbaumer, A., Ohtsuki, T., Shimazaki, T., Wada, T., Kirino, Y., Takai, K., Watanabe, K., Ramakrishnan, V., Suzuki, T.(2008) J Biological Chem 283: 18801
- PubMed: 18456657
- DOI: https://doi.org/10.1074/jbc.M800233200
- Primary Citation of Related Structures:
2VQE, 2VQF - PubMed Abstract:
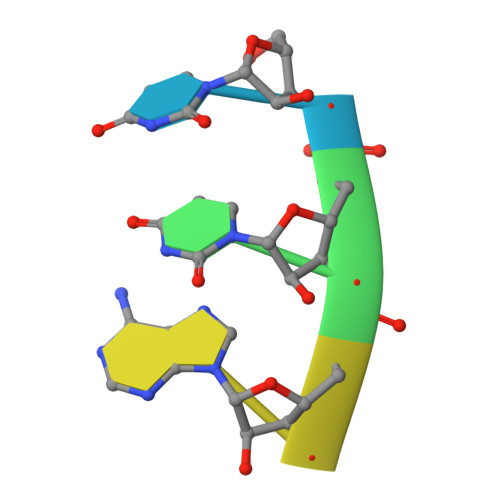
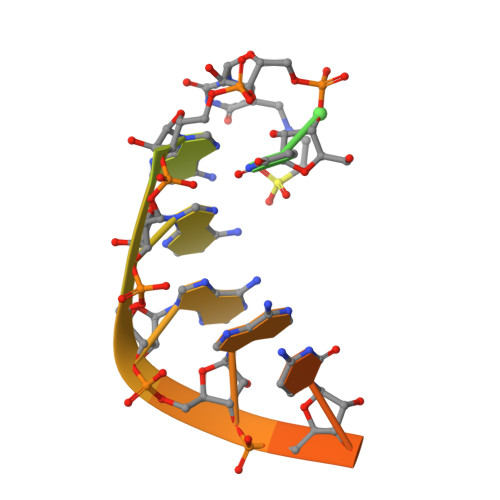
Post-transcriptional modifications at the first (wobble) position of the tRNA anticodon participate in precise decoding of the genetic code. To decode codons that end in a purine (R) (i.e. NNR), tRNAs frequently utilize 5-methyluridine derivatives (xm(5)U) at the wobble position. However, the functional properties of the C5-substituents of xm(5)U in codon recognition remain elusive. We previously found that mitochondrial tRNAs(Leu(UUR)) with pathogenic point mutations isolated from MELAS (mitochondrial myopathy, encephalopathy, lactic acidosis, and stroke-like episodes) patients lacked the 5-taurinomethyluridine (taum(5)U) modification and caused a decoding defect. Here, we constructed Escherichia coli tRNAs(Leu(UUR)) with or without xm(5)U modifications at the wobble position and measured their decoding activities in an in vitro translation as well as by A-site tRNA binding. In addition, the decoding properties of tRNA(Arg) lacking mnm(5)U modification in a knock-out strain of the modifying enzyme (DeltamnmE) were examined by pulse labeling using reporter constructs with consecutive AGR codons. Our results demonstrate that the xm(5)U modification plays a critical role in decoding NNG codons by stabilizing U.G pairing at the wobble position. Crystal structures of an anticodon stem-loop containing taum(5)U interacting with a UUA or UUG codon at the ribosomal A-site revealed that the taum(5)U.G base pair does not have classical U.G wobble geometry. These structures provide help to explain how the taum(5)U modification enables efficient decoding of UUG codons.
- Department of Chemistry and Biotechnology, Graduate School of Engineering, University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8656, Japan.
Organizational Affiliation: