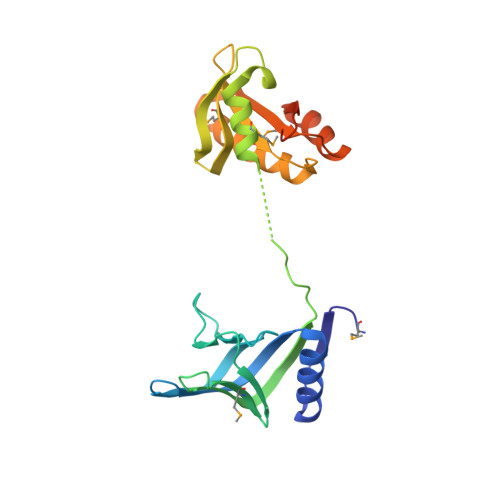
Structure and function of a novel endonuclease acting on branched DNA substrates.
Ren, B., Kuhn, J., Meslet-Cladiere, L., Briffotaux, J., Norais, C., Lavigne, R., Flament, D., Ladenstein, R., Myllykallio, H.(2009) EMBO J 28: 2479-2489
- PubMed: 19609302
- DOI: https://doi.org/10.1038/emboj.2009.192
- Primary Citation of Related Structures:
2VLD - PubMed Abstract:
We show that Pyrococcus abyssi PAB2263 (dubbed NucS (nuclease for ss DNA) is a novel archaeal endonuclease that interacts with the replication clamp PCNA. Structural determination of P. abyssi NucS revealed a two-domain dumbbell-like structure that in overall does not resemble any known protein structure. Biochemical and structural studies indicate that NucS orthologues use a non-catalytic ssDNA-binding domain to regulate the cleavage activity at another site, thus resulting into the specific cleavage at double-stranded DNA (dsDNA)/ssDNA junctions on branched DNA substrates. Both 3' and 5' extremities of the ssDNA can be cleaved at the nuclease channel that is too narrow to accommodate duplex DNA. Altogether, our data suggest that NucS proteins constitute a new family of structure-specific DNA endonucleases that are widely distributed in archaea and in bacteria, including Mycobacterium tuberculosis.
- Center for Structural Biochemistry, Karolinska Institutet, NOVUM, Huddinge, Sweden.
Organizational Affiliation: