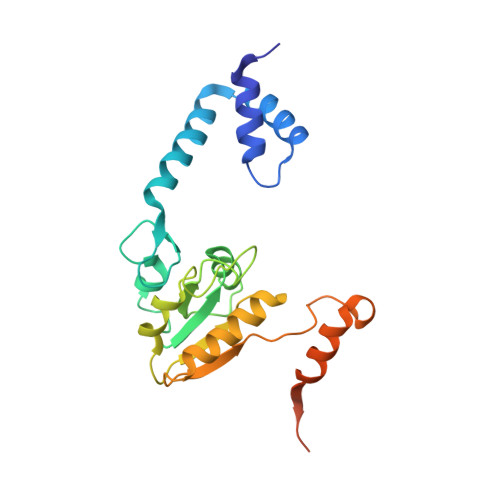
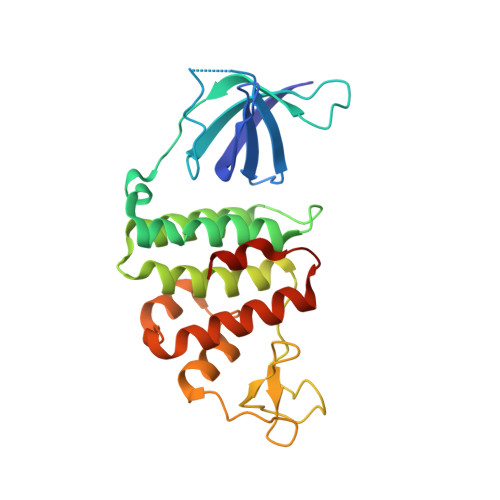
Crystal Structure and Mutational Study of Recor Provide Insight Into its Mode of DNA Binding.
Timmins, J., Leiros, I., Mcsweeney, S.(2007) EMBO J 26: 3260
- PubMed: 17581636
- DOI: https://doi.org/10.1038/sj.emboj.7601760
- Primary Citation of Related Structures:
2V1C - PubMed Abstract:
The crystal structure of the complex formed between Deinococcus radiodurans RecR and RecO (drRecOR) has been determined. In accordance with previous biochemical characterisation, the drRecOR complex displays a RecR:RecO molecular ratio of 2:1. The biologically relevant drRecOR entity consists of a heterohexamer in the form of two drRecO molecules positioned on either side of the tetrameric ring of drRecR, with their OB (oligonucleotide/oligosaccharide-binding) domains pointing towards the interior of the ring. Mutagenesis studies validated the protein-protein interactions observed in the crystal structure and allowed mapping of the residues in the drRecOR complex required for DNA binding. Furthermore, the preferred DNA substrate of drRecOR was identified as being 3'-overhanging DNA, as encountered at ssDNA-dsDNA junctions. Together these results suggest a possible mechanism for drRecOR recognition of stalled replication forks.
- Macromolecular Crystallography Group, European Synchrotron Radiation Facility, Grenoble-CEDEX 9, France.
Organizational Affiliation: