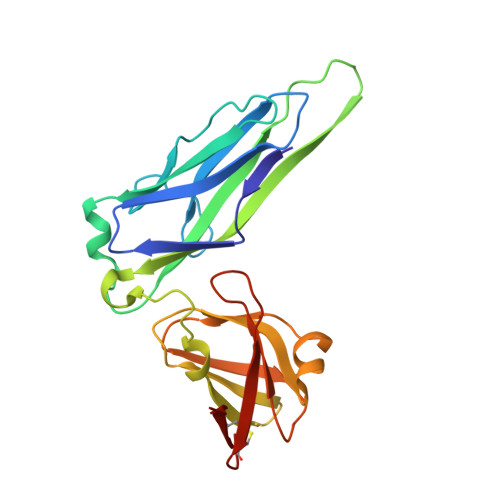
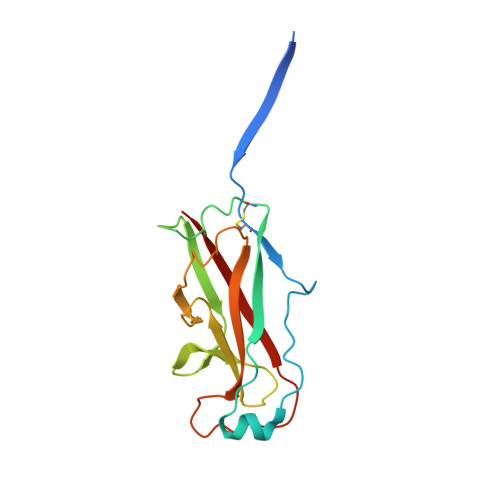
Crystal Structure of the P Pilus Rod Subunit Papa.
Verger, D., Bullitt, E., Hultgren, S.J., Waksman, G.(2007) PLoS Pathog 3: E73
- PubMed: 17511517
- DOI: https://doi.org/10.1371/journal.ppat.0030073
- Primary Citation of Related Structures:
2UY6, 2UY7 - PubMed Abstract:
P pili are important adhesive fibres involved in kidney infection by uropathogenic Escherichia coli strains. P pili are assembled by the conserved chaperone-usher pathway, which involves the PapD chaperone and the PapC usher. During pilus assembly, subunits are incorporated into the growing fiber via the donor-strand exchange (DSE) mechanism, whereby the chaperone's G1 beta-strand that complements the incomplete immunoglobulin-fold of each subunit is displaced by the N-terminal extension (Nte) of an incoming subunit. P pili comprise a helical rod, a tip fibrillum, and an adhesin at the distal end. PapA is the rod subunit and is assembled into a superhelical right-handed structure. Here, we have solved the structure of a ternary complex of PapD bound to PapA through donor-strand complementation, itself bound to another PapA subunit through DSE. This structure provides insight into the structural basis of the DSE reaction involving this important pilus subunit. Using gel filtration chromatography and electron microscopy on a number of PapA Nte mutants, we establish that PapA differs in its mode of assembly compared with other Pap subunits, involving a much larger Nte that encompasses not only the DSE region of the Nte but also the region N-terminal to it.
- Institute of Structural Molecular Biology, University College London and Birkbeck College, London, United Kingdom.
Organizational Affiliation: